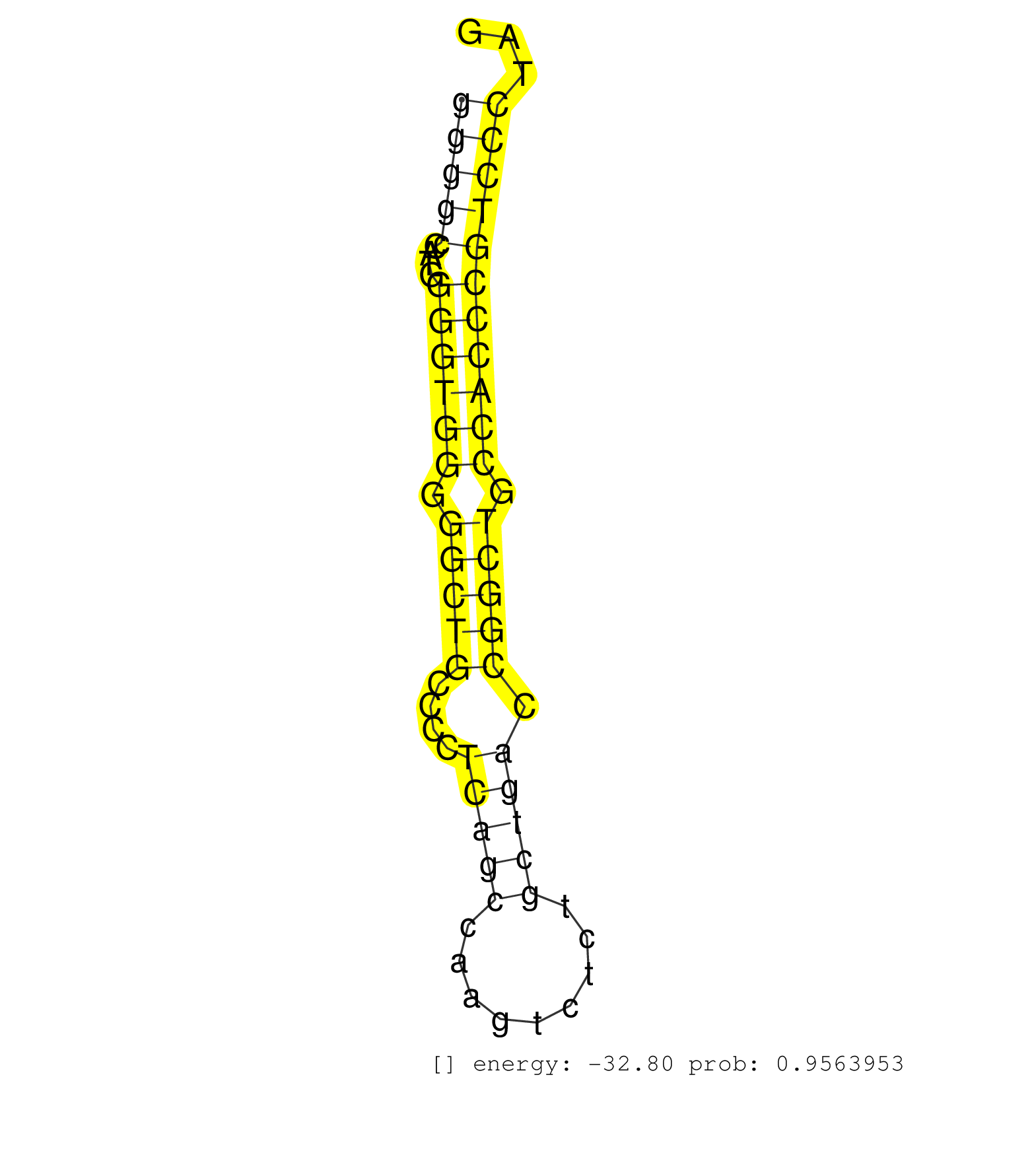

| Gene: Mfap4 | ID: uc007jhm.1_intron_3_0_chr11_61300509_f | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(13) TESTES |
| GAGCGACTACAAGCTGGGCTTTGGCCGTGCTGACGGGGAGTACTGGCTGGGTAAGGTGGGGCCATGGGGTGGGGGCTGCCCCTCAGCCAAGTCTCTGCTGACCGGCTGCCACCCGTCCCTAGGGCTGCAGAACCTGCACCTCCTGACACTGAAGCAGAAGTATGAGCTGCGC .........................................................(((((....((((((.(((((....(((((.........))))).))))).)))))))))))..................................................... .........................................................58..............................................................122................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................ACCGGCTGCCACCCGTCCCTAGt................................................. | 23 | t | 6.00 | 0.00 | - | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGACACTGAAGCAGAAGTATGA....... | 22 | 1 | 5.00 | 5.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTAGt................................................. | 22 | t | 5.00 | 1.00 | - | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................CATGGGGTGGGGGCTGCCCCTC........................................................................................ | 22 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AAGTCTCTGCTGACCGGC.................................................................. | 18 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTAGa................................................. | 22 | a | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................AACCTGCACCTCCTGACACTGAAGCAGA.............. | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGACCGGCTGCCACCCGTCCCTAGa................................................. | 25 | a | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................CTCCTGACACTGAAGCAGAA............. | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CTACAAGCTGGGCTTTGGCCGTGCTGACG......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................TCCTGACACTGAAGCAGAAGTATGAGC..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................CATGGGGTGGGGGCTGCCCCTat....................................................................................... | 23 | at | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................CTTTGGCCGTGCTGACG......................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTcgt................................................. | 22 | cgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGCACCTCCTGACACTGAAG.................. | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................CATGGGGTGGGGGCTGCCCCTCAattc................................................................................... | 27 | attc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CAGAACCTGCACCTCCTGACACTGAAGCAGAAGT........... | 34 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTAGtat............................................... | 24 | tat | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCCTGACACTGAAGCAGA.............. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGACACTGAAGCAGAAGTATGAGCcga.. | 27 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................AGGGCTGCAGAACCTGCACC................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTAGta................................................ | 23 | ta | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CGGCTGCCACCCGTCCCTAG.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................ACGGGGAGTACTGGCTGGGTA....................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................CTCCTGACACTGAAGCAGAAGTAT......... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AGAACCTGCACCTCCTGACAC....................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................TTTGGCCGTGCTGACGGGGA..................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGCAGAACCTGCACCTCCTGAC......................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCCTGACACTGAAGCAGAAGTATGA....... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACCGGCTGCCACCCGTCCCTAGttaa.............................................. | 26 | ttaa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CAGAACCTGCACCTCCTGACACTGAAGCA................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................ACCTGCACCTCCTGACACTGAAGCA................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTggt................................................. | 22 | ggt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CCGGCTGCCACCCGTCCCTAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CAGAACCTGCACCTCCTGACACTGAAGC................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCCTGACACTGAAGCAGAAGTATGAG...... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCCTGACACTGAAGCAGAAG............ | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGACACTGAAGCAGAAGT........... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................CTGACACTGAAGCAGAAGT........... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CGGCTGCCACCCGTCCCTAGt................................................. | 21 | t | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| GAGCGACTACAAGCTGGGCTTTGGCCGTGCTGACGGGGAGTACTGGCTGGGTAAGGTGGGGCCATGGGGTGGGGGCTGCCCCTCAGCCAAGTCTCTGCTGACCGGCTGCCACCCGTCCCTAGGGCTGCAGAACCTGCACCTCCTGACACTGAAGCAGAAGTATGAGCTGCGC .........................................................(((((....((((((.(((((....(((((.........))))).))))).)))))))))))..................................................... .........................................................58..............................................................122................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|