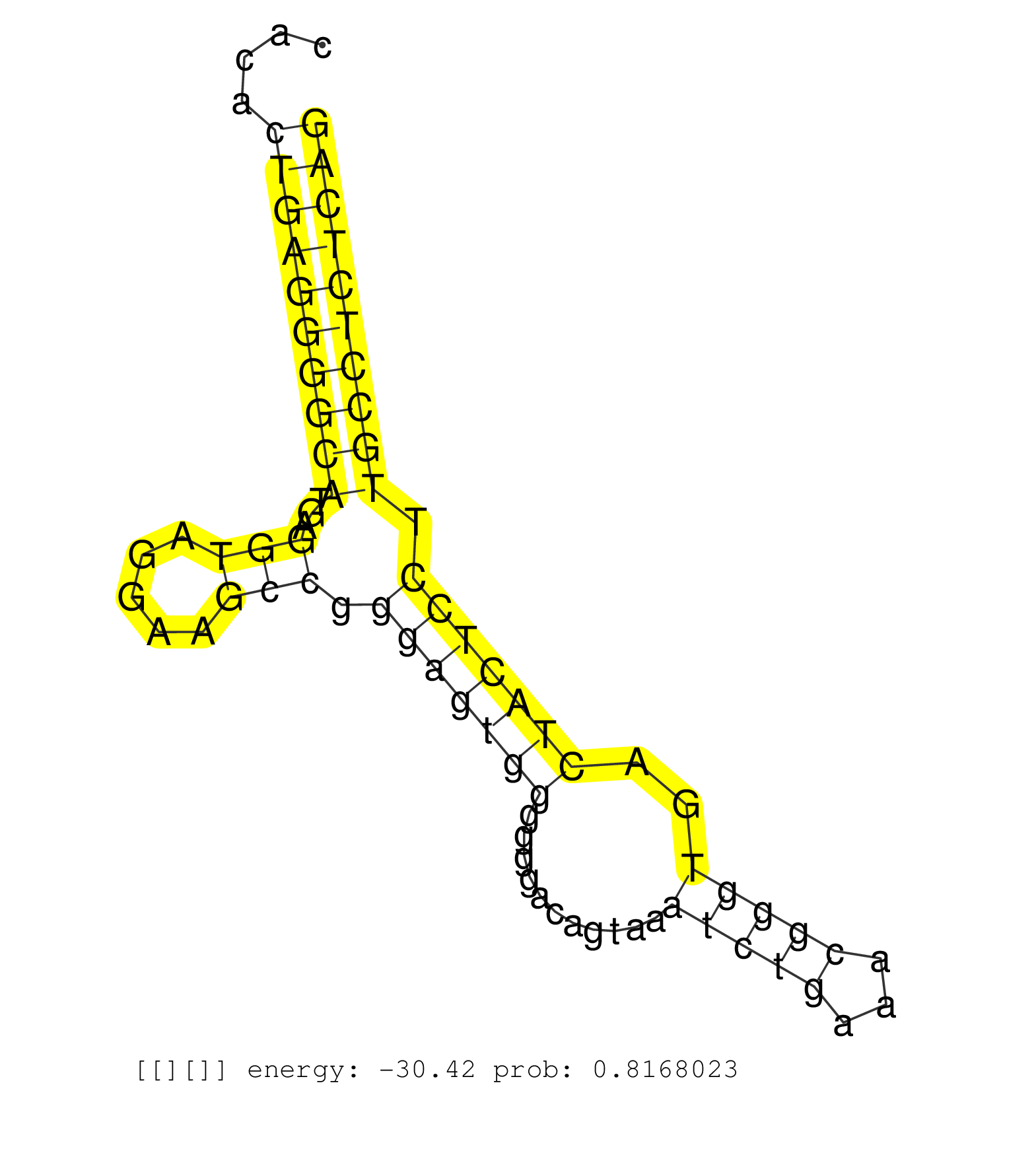
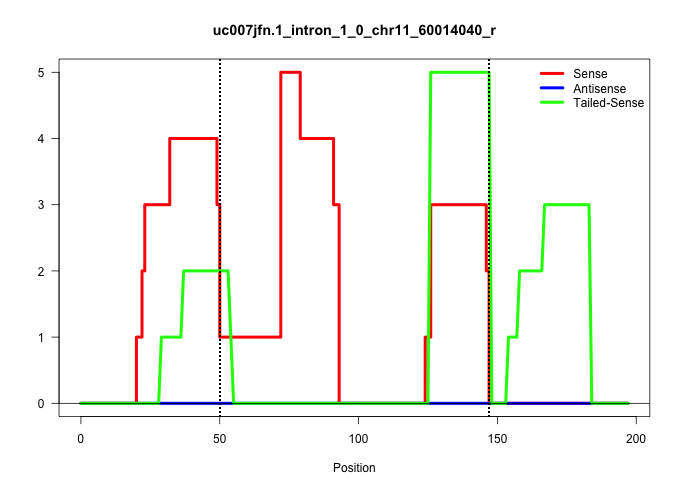
| Gene: Srebf1 | ID: uc007jfn.1_intron_1_0_chr11_60014040_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(17) TESTES |
| TGAGCAGCCTGCGGCGGTTGGCACAGAGCTTCCGGCCTGCTATGAGGAGGGTGAGTCCCACTGTTCTCACACTGAGGGGCATGAGGTAGGAAGCCGGGAGTGGGGGGACAGTAAATCTGAAACGGGTGACTACTCCTTGCCTCTCAGGTATTCCTACATGAGGCCACAGCTCGGCTGATGGCAGGAGCAAGTCCTGC .......................................................................((((((((((...(((.....))).(((((((...........(((((...)))))..))))))).)))))))))).................................................. ...................................................................68.............................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TGACTACTCCTTGCCTCTCAGa................................................. | 22 | a | 4.00 | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGACTACTCCTTGCCTCTCAGt................................................. | 22 | t | 4.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - |
| ........................................................................TGAGGGGCATGAGGTAGGAAG........................................................................................................ | 21 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................TGACTACTCCTTGCCTCTCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ..........................................................................................................................................................TACATGAGGCCACAGCTCGGCTGATGGCAa............. | 30 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GGTGACTACTCCTTGCCTCTCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTGAGTCCCACTGTTCTCACACTGAGGGG...................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TGAGGCCACAGCTCGGCTGATGGCAt............. | 26 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGCTATGAGGAGGGgtt............................................................................................................................................... | 17 | gtt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................CTGAGGGGCATGAGGTAGGAAG........................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................CACAGAGCTTCCGGCCTGCTATGAGGA..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................GAGCTTCCGGCCTGCTATGA........................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................TTCCGGCCTGCTATGAGGAGGGTatt.............................................................................................................................................. | 26 | att | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CAGAGCTTCCGGCCTGCTATGAGGAGG................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ACAGAGCTTCCGGCCTGCTATGAGGAG.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGAGGGGCATGAGGTAGGA.......................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CGGCCTGCTATGAGGAGG................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................GCACAGAGCTTCCGGCCTGCTATGAGGAGG................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................AGCTCGGCTGATGGCAt............. | 17 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGAGCAGCCTGCGGCGGTTGGCACAGAGCTTCCGGCCTGCTATGAGGAGGGTGAGTCCCACTGTTCTCACACTGAGGGGCATGAGGTAGGAAGCCGGGAGTGGGGGGACAGTAAATCTGAAACGGGTGACTACTCCTTGCCTCTCAGGTATTCCTACATGAGGCCACAGCTCGGCTGATGGCAGGAGCAAGTCCTGC .......................................................................((((((((((...(((.....))).(((((((...........(((((...)))))..))))))).)))))))))).................................................. ...................................................................68.............................................................................147................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................CTCCTTGCCTCTCAGGTATTCCTACA....................................... | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCCCACTGTTCTCAgctc................................................................................................................................ | 19 | gctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |