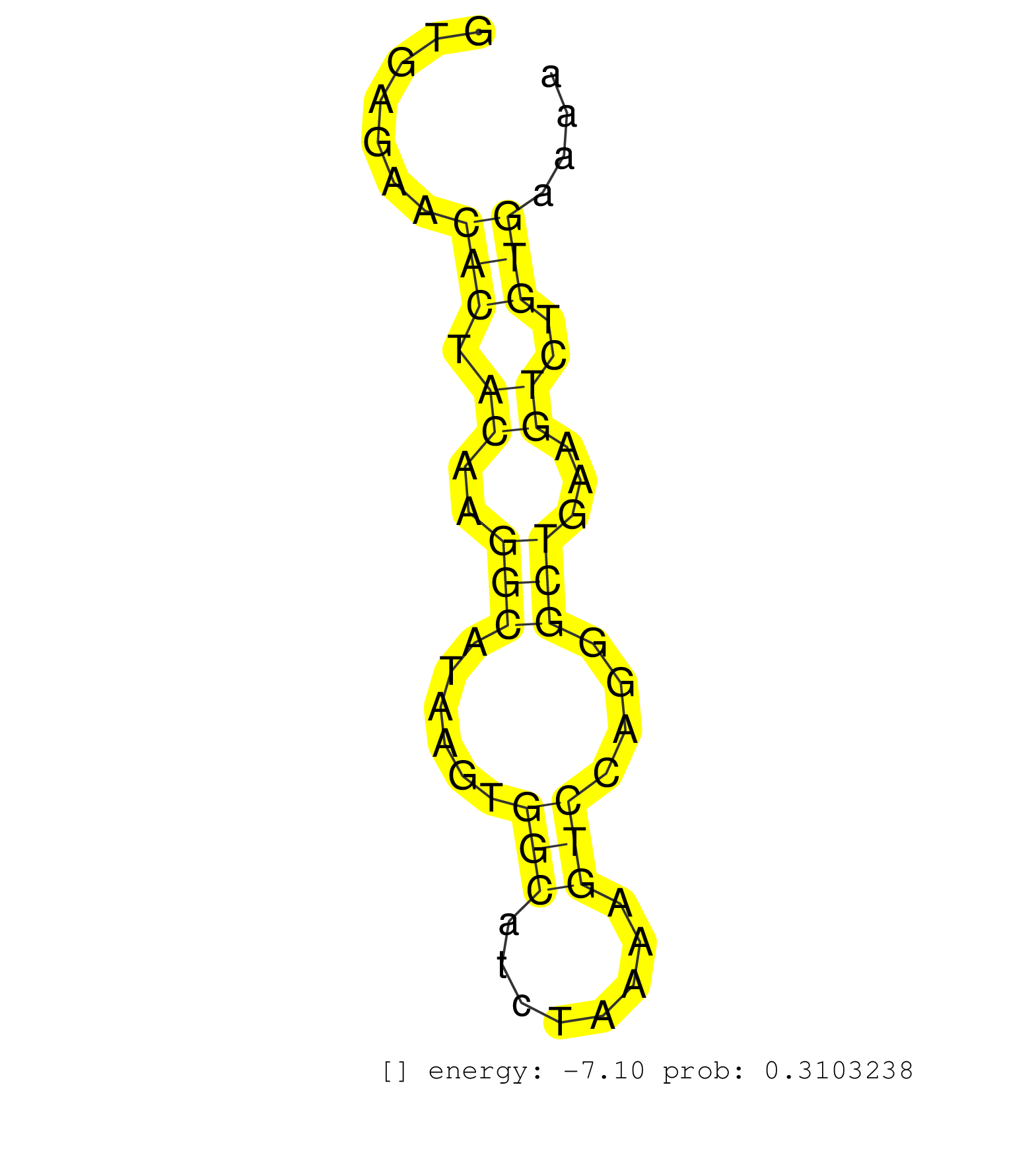

| Gene: RP23-180B18.4 | ID: uc007jem.1_intron_9_0_chr11_59558323_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| TCCTGGCAGATCAGAGCCTGAGATACTACAGGGATTCCGTGGCTGAGGAGGTGAGAACACTACAAGGCATAAGTGGCATCTAAAAGTCCAGGGCTGAAGTCTGTGAAAAGGTAGCCTACTGCCTTGGGTGTCATGGTTTCAGTCTACTGGGACAGTACATGCAGATGGGCCAGCTTTGATGGCAGAACTGTTGCCCTCCTCAGATATTATGGTCATTCTCTAAGGACTTGAGTTCGGTATCTTGATGCAG .........................................................(((.((..(((......(((........)))....)))...))..)))................................................................................................................................................. ..................................................51........................................................109........................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGAACACTACAAGGCATAAGTGGC............................................................................................................................................................................. | 27 | 1 | 65.00 | 65.00 | 25.00 | 20.00 | 9.00 | 6.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGG.............................................................................................................................................................................. | 26 | 1 | 12.00 | 12.00 | 6.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGGCAT........................................................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TCTAAGGACTTGAGTTCGGTATCTTGA..... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................TACTACAGGGATTCCGTGGCTGAGG.......................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGGCATC.......................................................................................................................................................................... | 30 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGGCA............................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TAAAAGTCCAGGGCTGAAGTCTGTG................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................AGGAGGTGAGAACACTACAAGG....................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGGCATa.......................................................................................................................................................................... | 30 | a | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GATTCCGTGGCTGAGGAGGcagc................................................................................................................................................................................................... | 23 | cagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGAACACTACAAGGCATAAGTGGC............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGgggc............................................................................................................................................................................. | 27 | gggc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAGATACTACAGGGATTCCGTGGC............................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................CCTGAGATACTACAGGGATTCCGTGGC............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGtc............................................................................................................................................................................. | 27 | tc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GATGGGCCAGCTTTGATGGCAGAACTGTTG......................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ATTCCGTGGCTGAGGA......................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGGGACAGTACATGCAGATGGGCCAGCTT.......................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CAGGGCTGAAGTCTGTGAAAAGGTAGC....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................GGATTCCGTGGCTGAGGA......................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TACATGCAGATGGGCCAGCTTTGAT...................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TACTGGGACAGTACATGCAGATGGt................................................................................. | 25 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGGTGTCATGGTTTCAGTCTACTGG.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TAAAAGTCCAGGGCTGAAGTCT.................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGGCAcc.......................................................................................................................................................................... | 30 | cc | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................ATGCAGATGGGCCAGCTTTGAT...................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................CAGTCTACTGGGACAGTACATGCAGATG................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTGAGAACACTACAAGGCA..................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TACTACAGGGATTCCGTGGCTGAGGAGG....................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TACATGCAGATGGGCCAGCTTTGATG..................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................AGGGATTCCGTGGCTGAGGAGGcag.................................................................................................................................................................................................... | 25 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...TGGCAGATCAGAGCCTGAGATACTACAGt.......................................................................................................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................TGGGACAGTACATGCAGATGGGCCAGC............................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TAAGGACTTGAGTTCGGTATCTTGAT.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TACTACAGGGATTCCGTGGCTGAGGA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................GGGATTCCGTGGCTGAGGAGGcag.................................................................................................................................................................................................... | 24 | cag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TTCCGTGGCTGAGGAGGcagc................................................................................................................................................................................................... | 21 | cagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACACTACAAGGCATAAGTGGCgtc.......................................................................................................................................................................... | 30 | gtc | 1.00 | 65.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGCAGATGGGCCAGCTTTGATGGCAG................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................CTGAGATACTACAGGGATTCCGTGGCTGAGGAG........................................................................................................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................AGATACTACAGGGATTCCGTGGCTG............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCTGGCAGATCAGAGCCTGAGATACTACAGGGATTCCGTGGCTGAGGAGGTGAGAACACTACAAGGCATAAGTGGCATCTAAAAGTCCAGGGCTGAAGTCTGTGAAAAGGTAGCCTACTGCCTTGGGTGTCATGGTTTCAGTCTACTGGGACAGTACATGCAGATGGGCCAGCTTTGATGGCAGAACTGTTGCCCTCCTCAGATATTATGGTCATTCTCTAAGGACTTGAGTTCGGTATCTTGATGCAG .........................................................(((.((..(((......(((........)))....)))...))..)))................................................................................................................................................. ..................................................51........................................................109........................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................GGTAGCCTACTGCCTccc.............................................................................................................................. | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TTGGGTGTCATGGaccc.................................................................................................................. | 17 | accc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |