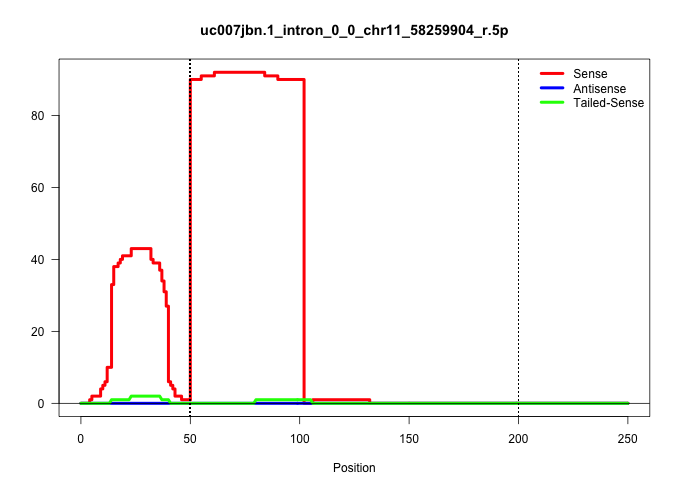
| Gene: 4930504O13Rik | ID: uc007jbn.1_intron_0_0_chr11_58259904_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(6) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(20) TESTES |
| TTCCACGGACAAGATAAAGTGTGTGGAGCTCTCCGGCATCATAAACACAGGTGCTTGAGAGAATGGACCCAAAAGCACGTTGAAAGGCGTAGAGCATAGTCACAAATGATAACTCCGGTGGCTGTTGCTGTTGTAATAGGCAGCTCAAGCCAAAAATGAGAGGTAATGTCTTCACGGTCCTAAAACATGTGCCTGGCTGCAGCGCAAGTCTGCACAGCTCCAAGGAGCAGCACAGGATGATCCCAGCACA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGCTTGAGAGAATGGACCCAAAAGCACGTTGAAAGGCGTAGAGCATAGTCA.................................................................................................................................................... | 52 | 1 | 90.00 | 90.00 | 90.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAAAGTGTGTGGAGCTCTCCGGCATC.................................................................................................................................................................................................................. | 26 | 1 | 19.00 | 19.00 | - | 6.00 | 4.00 | - | - | 3.00 | 1.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 |
| ............GATAAAGTGTGTGGAGCTCTCCGGCA.................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............GATAAAGTGTGTGGAGCTCT.......................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..............TAAAGTGTGTGGAGCTCTCCGGCATCATA............................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............AAAGTGTGTGGAGCTCTCCGGCATC.................................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AAAGTGTGTGGAGCTCTCCGGCAT................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGAGAGAATGGACCCAAAAGCACGTTGAA...................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........CAAGATAAAGTGTGTGGAGCTCTCCGGC..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........AGATAAAGTGTGTGGAGCTCTCCGGC..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........CAAGATAAAGTGTGTGGAGCTCTCCGG...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....ACGGACAAGATAAAGTGTGTGGAGCTCTC......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAAAGGCGTAGAGCATAGTCACgaa................................................................................................................................................ | 26 | gaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAAAGTGTGTGGAGCTCTCCGGCAT................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TGTGTGGAGCTCTCCGGCATCATAAAC............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAAAGTGTGTGGAGCTCTCCGGC..................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CGGACAAGATAAAGTGTGTGGAGCTCT.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................AGTGTGTGGAGCTCTCCGGCAT................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGGAGCTCTCCGGCATCt................................................................................................................................................................................................................. | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AAGATAAAGTGTGTGGAGCTCTCCGG...................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AATGGACCCAAAAGCACGTTGAAAGGCGT................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGGAGCTCTCCGGCATCA................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................TGATAACTCCGGTGGCTGTTGCTGTT...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................GTGTGTGGAGCTCTCCGGCA.................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AAAGTGTGTGGAGCTCTCCGGCATCAT................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAAAGTGTGTGGAGCTCTCCGGa..................................................................................................................................................................................................................... | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................TGGAGCTCTCCGGCATCATAAACACAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTCCACGGACAAGATAAAGTGTGTGGAGCTCTCCGGCATCATAAACACAGGTGCTTGAGAGAATGGACCCAAAAGCACGTTGAAAGGCGTAGAGCATAGTCACAAATGATAACTCCGGTGGCTGTTGCTGTTGTAATAGGCAGCTCAAGCCAAAAATGAGAGGTAATGTCTTCACGGTCCTAAAACATGTGCCTGGCTGCAGCGCAAGTCTGCACAGCTCCAAGGAGCAGCACAGGATGATCCCAGCACA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|