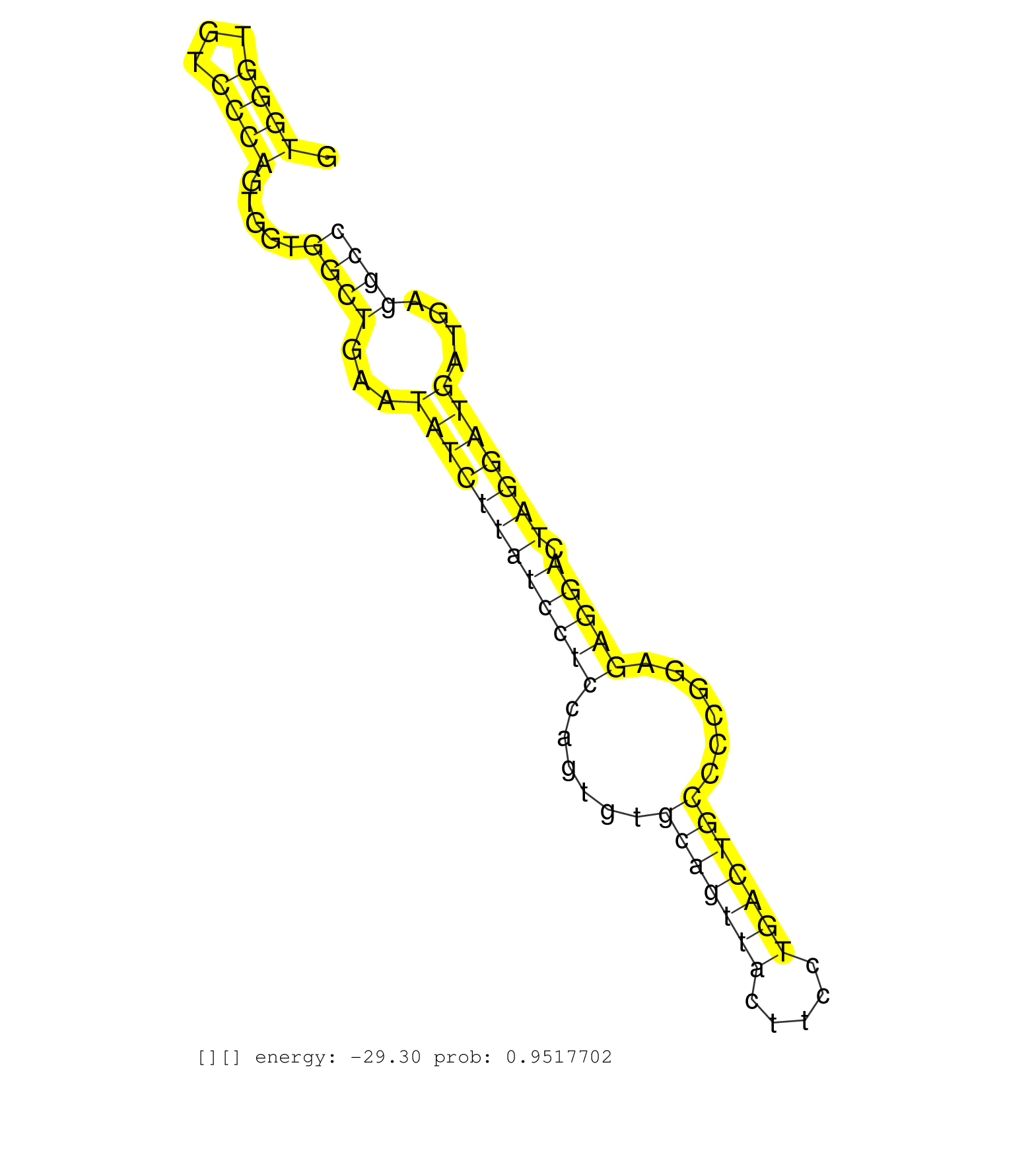

| Gene: Larp1 | ID: uc007jag.1_intron_5_0_chr11_57861372_f | SPECIES: mm9 |
 |
 |
|
(9) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(22) TESTES |
| TCTCAACTGCCCAGAGTTTGTTCCCCGCCAGCACTACCAGAAAGAGACAGGTGGGTGTCCCAGTGGTGGCTGAATATCTTATCCTCCAGTGTGCAGTTACTTCCTGACTGCCCCGGAGAGGACTAGGATGATGAGGCCCCACTTCTTCTCCCTTCAGAGTCGGCACCTGGCTCTCCCCGTGCAGTCACCCCAGTGCCAACAAAAACA ...................................................((((...)))).....((((...((((((((((((......(((((((.....)))))))......))))).)))))))....))))..................................................................... ..................................................51.....................................................................................138................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATATC................................................................................................................................. | 28 | 1 | 11.00 | 11.00 | 4.00 | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATAT.................................................................................................................................. | 27 | 1 | 8.00 | 8.00 | - | 1.00 | 1.00 | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATA................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATATt................................................................................................................................. | 28 | t | 4.00 | 8.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................TGGGTGTCCCAGTGGTGGCTGAATATC................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGA...................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGACTGCCCCGGAGAGGACTAGGATGATGt......................................................................... | 30 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATATCT................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAAaat.................................................................................................................................. | 27 | aat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGaaaa......................................................................................................................................... | 20 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................TTGTTCCCCGCCAGCACTACCAGAAA.................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCCGCCAGCACTACCAGAAAGAGACAGag........................................................................................................................................................... | 29 | ag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATAac................................................................................................................................. | 28 | ac | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TCCCCGCCAGCACTACCAGAAAGAGACA.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CCCCGGAGAGGACTAGGATGATGAa........................................................................ | 25 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................TTCCTGACTGCCCCGGAGAGGACTAGGATG............................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGTGTCCCAGTGGTGGCTGAATATCTTt.............................................................................................................................. | 30 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CTGCCCCGGAGAGGACTAGGATGATGAGG....................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGCTGAATt................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCTGACTGCCCCGGAGAGG...................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................CTTCCTGACTGCCCCGGAGAGGACTAG................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCTGACTGCCCCGGAGAGGACTAGGATGATGAG........................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................TACTTCCTGACTGCCCCGGAGAGGA..................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGGGTGTCCCAGTGGTGGC......................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGACTGCCCCGGAGAGGACTAGGATG............................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCCAGTGTGCAGTTACTTCCTGACT.................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTCAACTGCCCAGAGTTTGTTCCCCGCCAGCACTACCAGAAAGAGACAGGTGGGTGTCCCAGTGGTGGCTGAATATCTTATCCTCCAGTGTGCAGTTACTTCCTGACTGCCCCGGAGAGGACTAGGATGATGAGGCCCCACTTCTTCTCCCTTCAGAGTCGGCACCTGGCTCTCCCCGTGCAGTCACCCCAGTGCCAACAAAAACA ...................................................((((...)))).....((((...((((((((((((......(((((((.....)))))))......))))).)))))))....))))..................................................................... ..................................................51.....................................................................................138................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......CCAGAGTTTGTTCCCtgga...................................................................................................................................................................................... | 19 | tgga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |