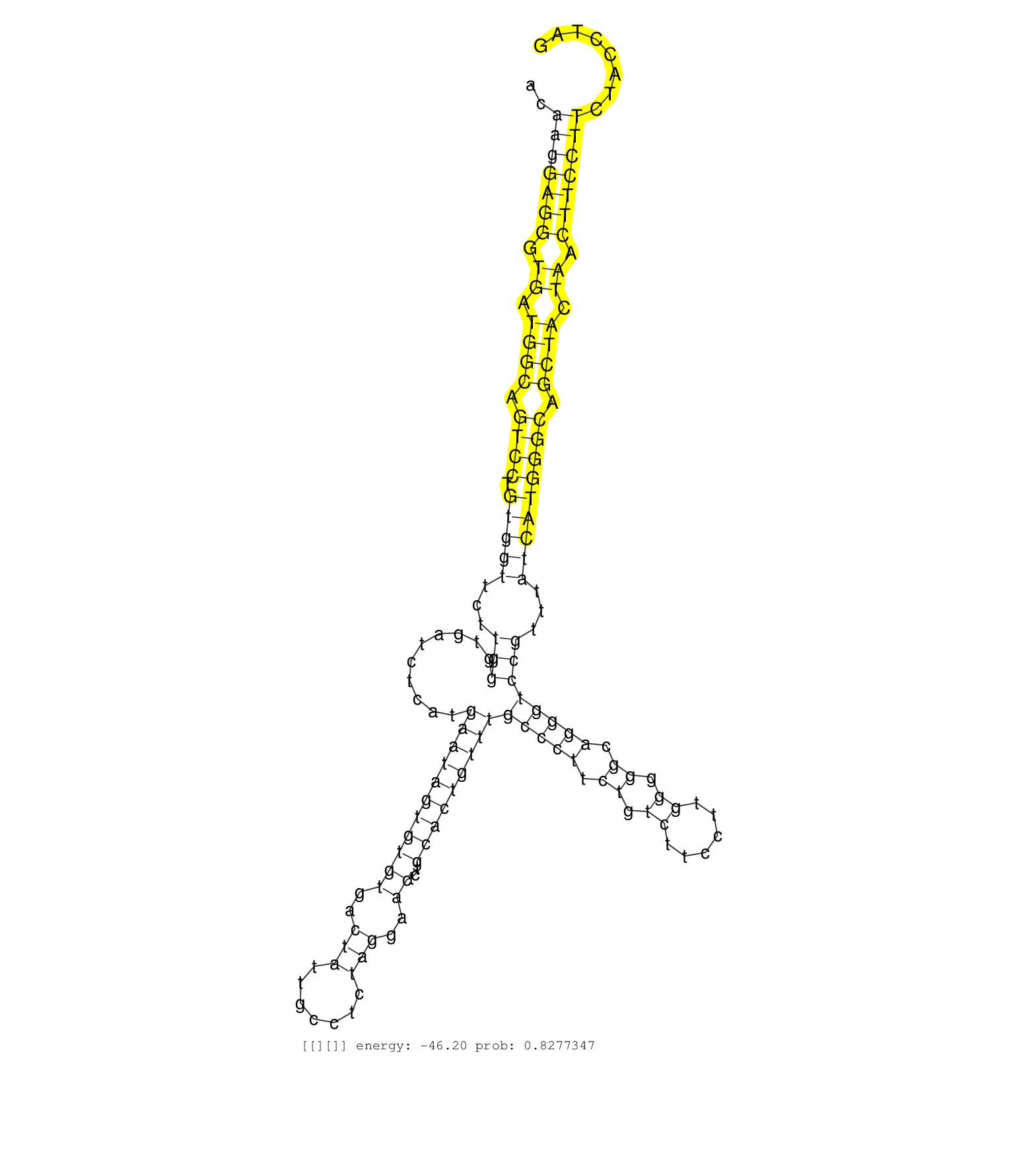
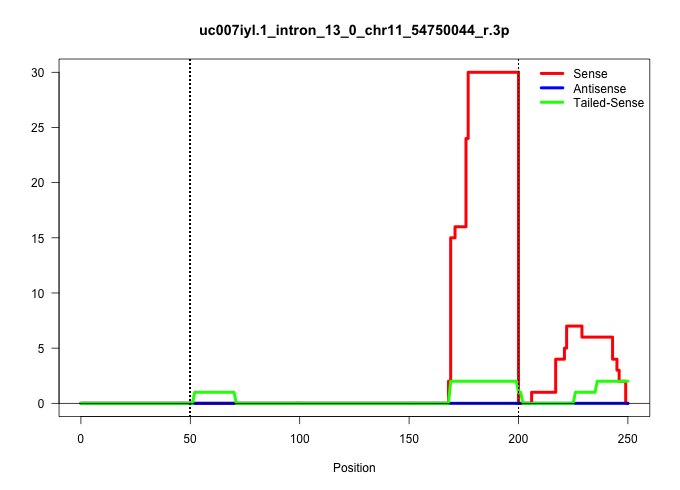
| Gene: Tnip1 | ID: uc007iyl.1_intron_13_0_chr11_54750044_r.3p | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(26) TESTES |
| CTGTTTCACTGATAAATAATGGCCATGGAGGGGTATGACCTCAGATCACAAGGAGGGTGATGGCAGTCCTGTGGTTCTTGGGTGATCTCATGAATAGTGTGTGACTATTGCCTCTAGGAACTCTGCACTGTTTGCCCTTCTGTCTTCCTTGGGGGCAGGGTCCGTTTATCATGGGCAGCTACTAACTTCCTTCTACCTAGGGCGCCTCCTCGGACTTTGAAGTGGTCCCTACTGAGGAGCAGAATTCACC .................................................(((((((.((.((((.((((.(((((...(((..........(((((((((((..(((.......)))..))...)))))))))(((((.((.((......)).)).))))))))...))))))))).)))).)).))))))).......................................................... ...............................................48......................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................CATGGGCAGCTACTAACTTCCTTCTACCTAG.................................................. | 31 | 1 | 16.00 | 16.00 | 1.00 | - | 1.00 | - | 2.00 | 2.00 | 2.00 | - | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - |
| .................................................................................................................................................................................GCTACTAACTTCCTTCTACCTAG.................................................. | 23 | 1 | 10.00 | 10.00 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AGCTACTAACTTCCTTCTACCTAG.................................................. | 24 | 1 | 10.00 | 10.00 | 1.00 | 1.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................................................GGAGCAGAATTCACCaagc | 19 | aagc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................................TGAAGTGGTCCCTACTGAGGAGCAGA....... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGTCCCTACTGAGGAGCAGAATTCAC. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TCATGGGCAGCTACTAACTTCCTTCTACCTAG.................................................. | 32 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................CATGGGCAGCTACTAACTTCCTTCTACCTAGaa................................................ | 33 | aa | 1.00 | 16.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TTTGAAGTGGTCCCTACTG................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................GAGCAGAATTCACCaagc | 18 | aagc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GAGGGTGATGGCAGTaatc................................................................................................................................................................................... | 19 | aatc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................GTGGTCCCTACTGAGGAGCAGAATT.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGAAGTGGTCCCTACTGAGGAGCAGAAT..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................CCCTACTGAGGAGCAGAATTCACCcgt | 27 | cgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCCTCGGACTTTGAAGTGGTCCC..................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................CAGCTACTAACTTCCTTCTACCTAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGGCAGCTACTAACTTCCTTCTACCTAG.................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CATGGGCAGCTACTAACTTCCTTCTACCcag.................................................. | 31 | cag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGTTTCACTGATAAATAATGGCCATGGAGGGGTATGACCTCAGATCACAAGGAGGGTGATGGCAGTCCTGTGGTTCTTGGGTGATCTCATGAATAGTGTGTGACTATTGCCTCTAGGAACTCTGCACTGTTTGCCCTTCTGTCTTCCTTGGGGGCAGGGTCCGTTTATCATGGGCAGCTACTAACTTCCTTCTACCTAGGGCGCCTCCTCGGACTTTGAAGTGGTCCCTACTGAGGAGCAGAATTCACC .................................................(((((((.((.((((.((((.(((((...(((..........(((((((((((..(((.......)))..))...)))))))))(((((.((.((......)).)).))))))))...))))))))).)))).)).))))))).......................................................... ...............................................48......................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|