
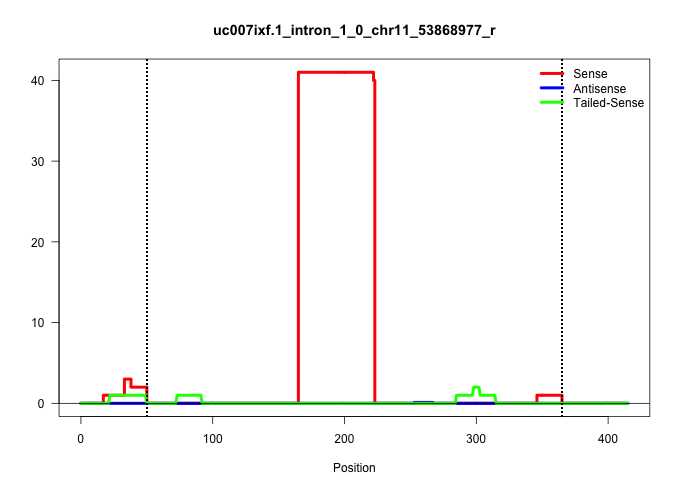
| Gene: Pdlim4 | ID: uc007ixf.1_intron_1_0_chr11_53868977_r | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(13) TESTES |
| GTCAGGATCCTTCCGCTACCTGCAGGGCATGCTAGAGGCTGGCGAGGGCGGTAAGACTCCCAACACCTTACCTCCCCTGCCTCCCCTCCCCCTACCGTGCCCAGTTTACCCTTCACCTTCCAACACCACCCAATCCACACCTTCTAGCCATCCACCCCCTCTTCAAATCCCTCTGTGCCTTACTGTGACTGTGCTCTCACCCTCCCGATTCCCACTCCACCACCCACGTGACTCTCCTTTACCTCATACAAACCAAGAACTTGTCCTCTAAGCTTCTGGTCATCGTGTTCCAGAAGCAACATCCTGCTGAAGTAGGGTTCTTATCTCACCTACCCCATCCATACCACCTGCCCTCTGGATTTCAGGGGATCGGCCTGGGTCTGGTGGTCCCCGGAACCTCAAACCAGCAGCCAGC .............................................................................................................................................................................(((..(((.(((..((.(((....))).....((((..((.......(((....))).........................((((((((.....)))).))))))..)))).))..))))))..))).................................................................................................................. ................................................................................................................................................................161...............................................................................................................................................307.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................AATCCCTCTGTGCCTTACTGTGACTGTGCTCTCACCCTCCCGATTCCCACTC...................................................................................................................................................................................................... | 52 | 1 | 41.00 | 41.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGAGGCTGGCGAGGGCG............................................................................................................................................................................................................................................................................................................................................................................. | 17 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................TGTTCCAGAAGCAAaaaa................................................................................................................ | 18 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................ATCGGCCTGGGTCTGGTGGT........................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................CCCCTGCCTCCCCTCCgcg................................................................................................................................................................................................................................................................................................................................... | 19 | gcg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................CCTGCCCTCTGGATTTCAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................TCCCCTGCCTCCCCTCCgc.................................................................................................................................................................................................................................................................................................................................... | 19 | gc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................ATCGGCCTGGGTCTGGTGGTCCCCGGAACCT................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................CCGGAACCTCAAACCAGCAGCCA.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................CAGGGCATGCTAGAGGCTGGCGAGGGgg............................................................................................................................................................................................................................................................................................................................................................................. | 28 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................................................................................................................................................ACATCCTGCTGAAtgga.................................................................................................... | 17 | tgga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................CCCCTGCCTCCCCTCCgcgc.................................................................................................................................................................................................................................................................................................................................. | 20 | gcgc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....GGATCCTTCCGCTACCTGCAGG..................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................ACCTGCAGGGCATGCTAGAGG......................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| GTCAGGATCCTTCCGCTACCTGCAGGGCATGCTAGAGGCTGGCGAGGGCGGTAAGACTCCCAACACCTTACCTCCCCTGCCTCCCCTCCCCCTACCGTGCCCAGTTTACCCTTCACCTTCCAACACCACCCAATCCACACCTTCTAGCCATCCACCCCCTCTTCAAATCCCTCTGTGCCTTACTGTGACTGTGCTCTCACCCTCCCGATTCCCACTCCACCACCCACGTGACTCTCCTTTACCTCATACAAACCAAGAACTTGTCCTCTAAGCTTCTGGTCATCGTGTTCCAGAAGCAACATCCTGCTGAAGTAGGGTTCTTATCTCACCTACCCCATCCATACCACCTGCCCTCTGGATTTCAGGGGATCGGCCTGGGTCTGGTGGTCCCCGGAACCTCAAACCAGCAGCCAGC .............................................................................................................................................................................(((..(((.(((..((.(((....))).....((((..((.......(((....))).........................((((((((.....)))).))))))..)))).))..))))))..))).................................................................................................................. ................................................................................................................................................................161...............................................................................................................................................307.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................CATACAAACCAAGAACTTGTCCTCTA................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CCAACACCACCCAATcttg......................................................................................................................................................................................................................................................................................... | 19 | cttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................................................CAAGAACTTGTCCTC................................................................................................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - |