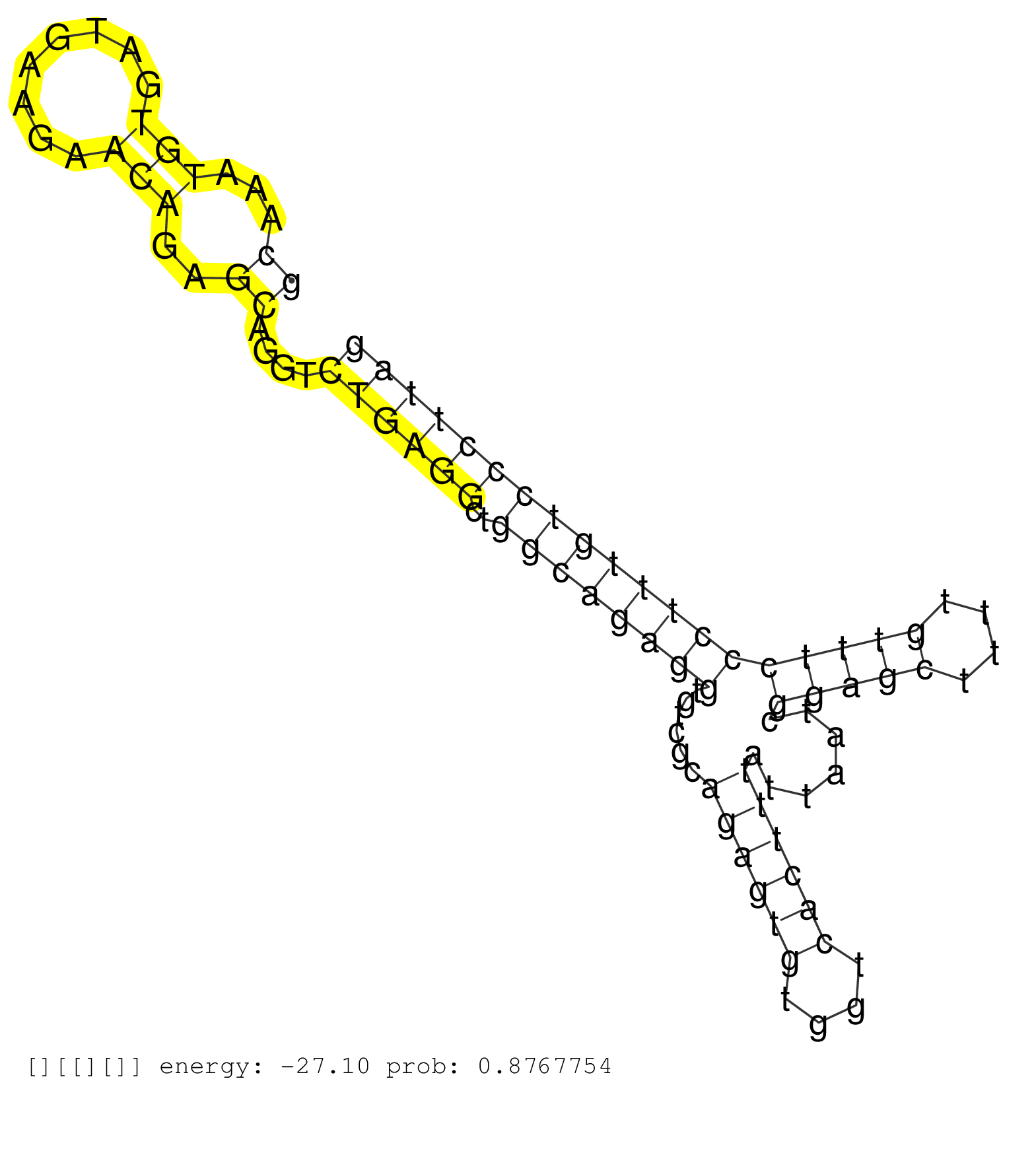

| Gene: Zcchc10 | ID: uc007ivt.1_intron_1_0_chr11_53140930_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| ACGTGTGTTTTATCACAGAGTCGGCCACATGCACTGTGTACATCCTGTCCCGGGAGGAAAATGTTTGACTTGATTAGAGCAGGAAACCCTGTGCCCTTGAGCAAATGTGATGAAGAACAGAGCAGGTCTGAGGCTGGCAGAGGTGTCGCAGAGTGTGGTCACTTTATTAATCGGAGCTTTTGTTTCCCTTTGTCCCTTAGCATAGGAGAGACCAACATAGAGAAAAAGATCAAGAAGAAGAAGAGGTAGG ....................................................................................................((...(((........)))..))....((((((..((((((((......((((((....)))))).......(((((....))))))))))))))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................AAATGTGATGAAGAACAGAGCAGGTCTGAGG..................................................................................................................... | 31 | 1 | 34.00 | 34.00 | 10.00 | 8.00 | 4.00 | 3.00 | 3.00 | 3.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................................TAGAGAAAAAGATCAAGAAGAAGAAG....... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................CAAATGTGATGAAGAACAGAGCAGGTCTGAGG..................................................................................................................... | 32 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AAATGTGATGAAGAACAGAGCAGGTCTGAGGat................................................................................................................... | 33 | at | 2.00 | 34.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGGAGAGACCAACATAGAGAAAAAGATC................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................TAGAGAAAAAGATCAAGAAGAAGAAGt...... | 27 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................ATGTGATGAAGAACAGAGCAGGTCTGAG...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GAGAAAAAGATCAAGAAGAAGAAGAGGT... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AAATGTGATGAAGAACAGAGCAGGTCTGA....................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................AAAGATCAAGAAGAAGAAGAGGgatg | 26 | gatg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................ATAGGAGAGACCAACATAGAGAAAAAG...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................................................TAGAGAAAAAGATCAAGAAGAAGAAGAGG.... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TAGGAGAGACCAACATAGAGAAAAAGAT.................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AAATGTGATGAAGAACAGAGCAGGTCTGAGGC.................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AGGTCTGAGGCTGGCAGA............................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ACGTGTGTTTTATCACAGAGTCGGCCACATGCACTGTGTACATCCTGTCCCGGGAGGAAAATGTTTGACTTGATTAGAGCAGGAAACCCTGTGCCCTTGAGCAAATGTGATGAAGAACAGAGCAGGTCTGAGGCTGGCAGAGGTGTCGCAGAGTGTGGTCACTTTATTAATCGGAGCTTTTGTTTCCCTTTGTCCCTTAGCATAGGAGAGACCAACATAGAGAAAAAGATCAAGAAGAAGAAGAGGTAGG ....................................................................................................((...(((........)))..))....((((((..((((((((......((((((....)))))).......(((((....))))))))))))))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................GTCTGAGGCTGGCAGAaca............................................................................................................. | 19 | aca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................TGTCGCAGAGTGTtta.............................................................................................. | 16 | tta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |