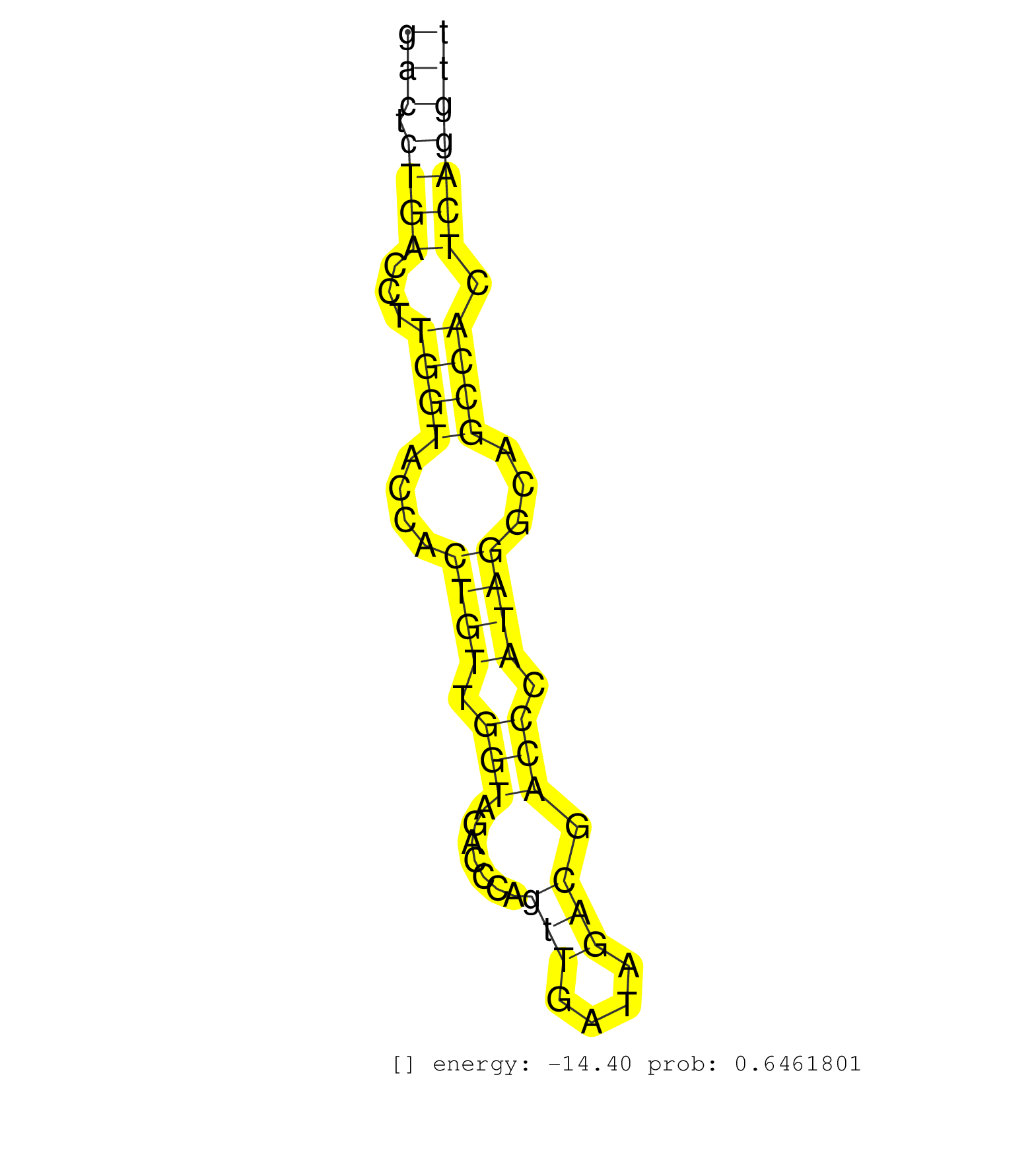
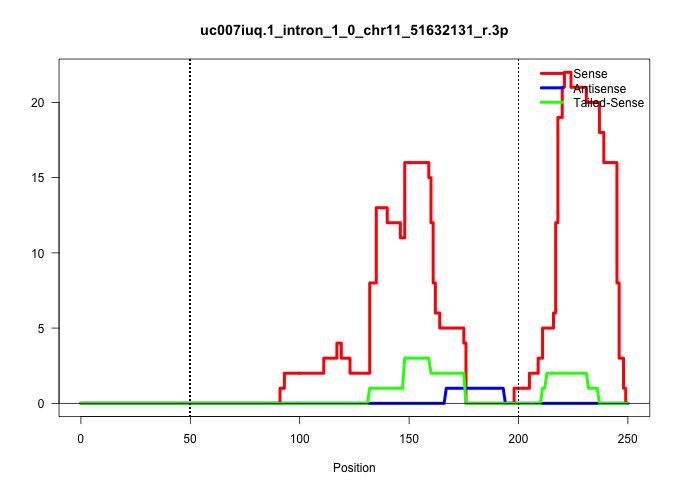
| Gene: Phf15 | ID: uc007iuq.1_intron_1_0_chr11_51632131_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| AGTCCAGTTCGTAGTGCACTTTGAAGTCTAGGTCCCACAAGGACTTATTATGCCTTCCTCTATCCTTCATAGAGCTGTGGGCTGCCCCCGCTGCCCCGGTTTCTGCCACACTGACTCTGACCTTGGTACCACTGTTGGTAGACCCAGTTGATAGACGACCCATAGGCAGCCACTCAGGTTTAACTCTTTCTCCAACACAGAGCCTTCTGGGAGGAGGTCAAAGGGCAAGAAGAATGATTCAAAAAGGAAA ................................................................................................................(((.((((...((((....((((.(((.......(((....))).))).))))...)))).)))))))...................................................................... ................................................................................................................113................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................CAAAGGGCAAGAAGAATGATTCAAAAA..... | 27 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................................TCAAAGGGCAAGAAGAATGATTCAAAAA..... | 28 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGATAGACGACCCATAGGCAGCCACTCA.......................................................................... | 28 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTTGGTAGACCCAGTTGATAGACGACC.......................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................AAGGGCAAGAAGAATGATTCAAAAAGGA.. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTTGGTAGACCCAGTTGATAGACGACCC......................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................TGATAGACGACCCATAGGCAGCCACTCt.......................................................................... | 28 | t | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGTAGACCCAGTTGATAGACGACCC......................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CAAAGGGCAAGAAGAATGATTCAAAAAG.... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TCAAAGGGCAAGAAGAATGATTCAAAAAG.... | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGTAGACCCAGTTGATAGACGACCCA........................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGACCTTGGTACCACTGTTGGTAGACCCA........................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCTGGGAGGAGGTCAAAGGGCAAGAA................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CAAAGGGCAAGAAGAATGATT........... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................AGAGCCTTCTGGGAGGAGGTCAAAGG.......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGATAGACGACCCATAGGCAGCCACTC........................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AGGGCAAGAAGAATGATTCAAAAAGGAA. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGGAGGTCAAAGGGCAAGAAGAATGg............. | 26 | g | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTTGGTAGACCCAGTTGATAGACGAC........................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GGAGGAGGTCAAAGGGCAAGAAGAATGA............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................CCCCGGTTTCTGCCACACTGACTCTGACCT............................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................TGACTCTGACCTTGGTACCACTGTTGGTA.............................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................GAGGTCAAAGGGCAAGAAa.................. | 19 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGGAGGTCAAAGGGCAAGAAGAATGA............. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTTGGTAGACCCAGTTGATAGACGACa.......................................................................................... | 28 | a | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGTAGACCCAGTTGATAGACGACCCATA...................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGGAGGTCAAAGGGCAAGAAGAATGATT........... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GTCAAAGGGCAAGAAGAATGATTCAAAAAG.... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGCCCCGGTTTCTGCCACACTGACTCTG................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| AGTCCAGTTCGTAGTGCACTTTGAAGTCTAGGTCCCACAAGGACTTATTATGCCTTCCTCTATCCTTCATAGAGCTGTGGGCTGCCCCCGCTGCCCCGGTTTCTGCCACACTGACTCTGACCTTGGTACCACTGTTGGTAGACCCAGTTGATAGACGACCCATAGGCAGCCACTCAGGTTTAACTCTTTCTCCAACACAGAGCCTTCTGGGAGGAGGTCAAAGGGCAAGAAGAATGATTCAAAAAGGAAA ................................................................................................................(((.((((...((((....((((.(((.......(((....))).))).))))...)))).)))))))...................................................................... ................................................................................................................113................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................AGCCACTCAGGTTTAACTCTTTCTCCA........................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................GCCACTCAGGTTTAACTCTTTCTCCA........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |