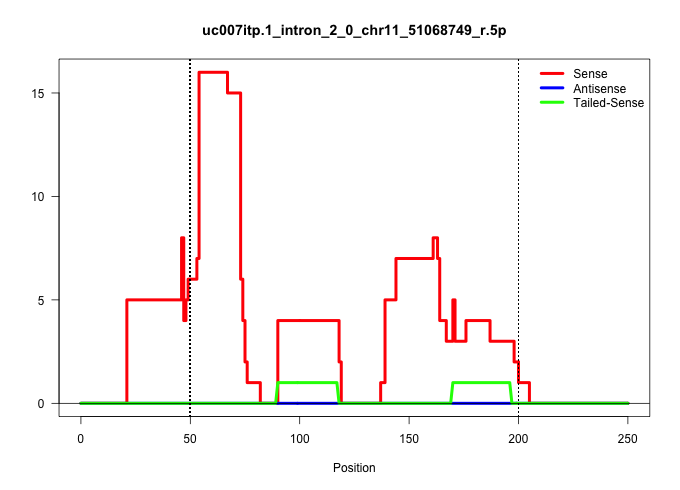
| Gene: BC049762 | ID: uc007itp.1_intron_2_0_chr11_51068749_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| GGAAGCTCATCGCACTCCATCTACCAGTCGCCCCTCAACCTCAGCTCCAGGTAGAGCCACCAGAGCGCAGAGTCCCCGAATGCTCCCAGCTAGGGCAGCTCAGTGACCAGGATATGGCTGTCACCTGGCGCTTCCGCTCTAAACTGGATTTGGAGAAGTTTACCTGAATATGCCTCTAAAAACAAGCGTAGCAGGTGCTTACCCTTCTCTTTCTCTTTGCCTTCCTCTCTCTCTTCCTCCTCGCCTTCTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................AGCCACCAGAGCGCAGAGT................................................................................................................................................................................. | 19 | 1 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TACCAGTCGCCCCTCAACCTCAGCTC........................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................TAAACTGGATTTGGAGAAGTTTACC...................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TAGGGCAGCTCAGTGACCAGGATATGGCT................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CCAGGTAGAGCCACCAGAGCGCAGAGTC................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................TGGATTTGGAGAAGTTTACCTGAATAT............................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TAGGGCAGCTCAGTGACCAGGATATGGC.................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCCTCTAAAAACAAGCGTAGCAGGTGC.................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GAGCCACCAGAGCGCAGAGTCCCCGAATG........................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTAGAGCCACCAGAGCG....................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................TACCAGTCGCCCCTCAACCTCAGCTCC.......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................ACCTGAATATGCCTCTAAAAACAAGC............................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCCTCTAAAAACAAGCGTAGCAGGTGCTT.................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TAGGGCAGCTCAGTGACCAGGATATGGt.................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTAGAGCCACCAGAGCGCAGAGTCCC.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAAACTGGATTTGGAGAAGTTTACCTGA................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................TAAAAACAAGCGTAGCAGGTGCTTACCCT............................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CCAGGTAGAGCCACCAGAGCGCAGAGTCC............................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TCTAAACTGGATTTGGAGAAGTTTAC....................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................AGGTAGAGCCACCAGAGCGCAGAGTCC............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................TGCCTCTAAAAACAAGCGTAGCAGGTt..................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GGAAGCTCATCGCACTCCATCTACCAGTCGCCCCTCAACCTCAGCTCCAGGTAGAGCCACCAGAGCGCAGAGTCCCCGAATGCTCCCAGCTAGGGCAGCTCAGTGACCAGGATATGGCTGTCACCTGGCGCTTCCGCTCTAAACTGGATTTGGAGAAGTTTACCTGAATATGCCTCTAAAAACAAGCGTAGCAGGTGCTTACCCTTCTCTTTCTCTTTGCCTTCCTCTCTCTCTTCCTCCTCGCCTTCTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TCTACCAGTCGaatc............................................................................................................................................................................................................................ | 15 | aatc | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |