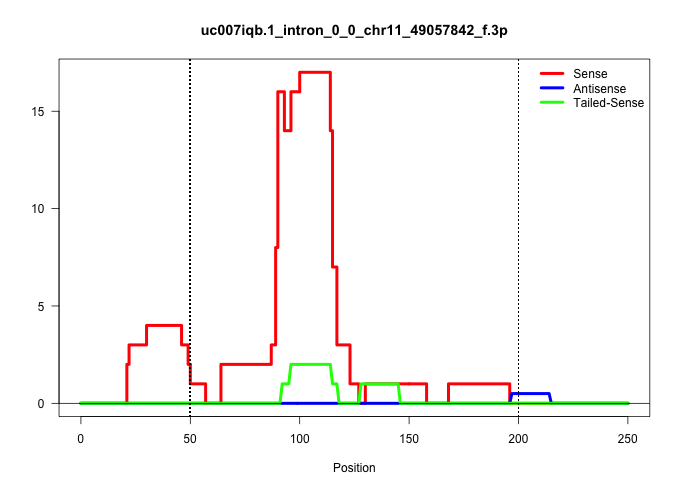
| Gene: Mgat1 | ID: uc007iqb.1_intron_0_0_chr11_49057842_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| AACACACATAAACACATATGCTCAAAATGATGGGGATACAGTATGTGCATGAAGGATGTACTTTTTATACAAATTGTGTGGCGTACTCAAGAGAGTTTGTTGACTGAACACGGTAAAGGAGTGTGGTGAATATGGAAGTCTTAGAAATCCACTGGGGCATGTATTAGATTAGAGTGTCATACTGTTTAAATGTTCCACAGGTTCCCACAGAGATCAGCCTCCCGGGCCATGTTGGTGTTAAAGCCTGTTT .....................................................................................(((....)))..((((.(((..((((.........))))))).)))).(((..((...))..)))..(((..(((...((((((...((((...)))))))))))))..)))..................................................... .....................................................................................86................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................TACAAATTGTGTGGCGTACTCAAGAG............................................................................................................................................................. | 26 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GAGAGTTTGTTGACTGAACACGGTAAA..................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACAGAGATCAGCCTCCCGGGCCATGTTGG............... | 29 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GAGAGTTTGTTGACTGAACACGGTA....................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| .........................................................................................AGAGAGTTTGTTGACTGAACACGGTA....................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TTATACAAATTGTGTGGCGTACTCAAGAG............................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTGTTGACTGAACACGGTAAAGGAGTG............................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AGAGAGTTTGTTGACTGAACACGGT........................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................CAAGAGAGTTTGTTGACTGAACACGGTA....................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTGTTGACTGAACACattc....................................................................................................................................... | 19 | attc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................TCAAAATGATGGGGATACAGTATGT............................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................GAGAGTTTGTTGACTGAACACGGT........................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TTAGAGTGTCATACTGTTTAAATGTTCC...................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................TGACTGAACACGGTAAAGGAGTGTGGT........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................TGGGGATACAGTATGTGCATGAAGGAT................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TATGGAAGTCTTAGAAATCCACTGGGGC............................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................CAAAATGATGGGGATACAGTATGTGCAT........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AATATGGAAGTCTTcatc........................................................................................................ | 18 | catc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................GAGTTTGTTGACTGAACACGGTtccc.................................................................................................................................... | 26 | tccc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TCAAAATGATGGGGATACAGTATGTGCA......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| AACACACATAAACACATATGCTCAAAATGATGGGGATACAGTATGTGCATGAAGGATGTACTTTTTATACAAATTGTGTGGCGTACTCAAGAGAGTTTGTTGACTGAACACGGTAAAGGAGTGTGGTGAATATGGAAGTCTTAGAAATCCACTGGGGCATGTATTAGATTAGAGTGTCATACTGTTTAAATGTTCCACAGGTTCCCACAGAGATCAGCCTCCCGGGCCATGTTGGTGTTAAAGCCTGTTT .....................................................................................(((....)))..((((.(((..((((.........))))))).)))).(((..((...))..)))..(((..(((...((((((...((((...)))))))))))))..)))..................................................... .....................................................................................86................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................CAGGTTCCCACAGAGATC................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |