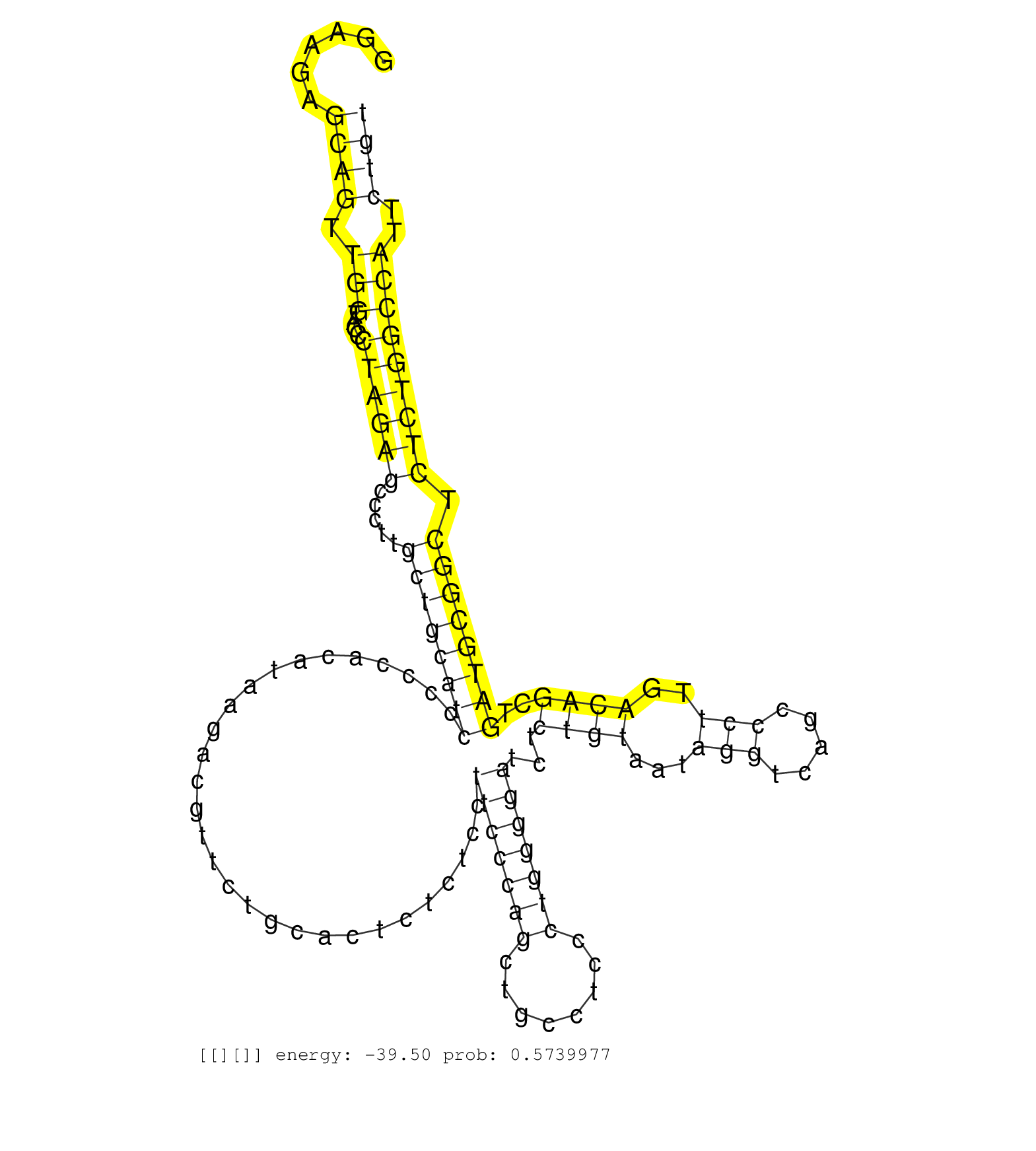

| Gene: Wwc1 | ID: uc007ili.1_intron_8_0_chr11_35680930_r.3p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(9) TESTES |
| TAGGAAAAAAAGAAACAAAATAACAACAAAAAATTCAGAGAGCCATGGCAGGAAGAGCAGTTGGTACCCTAGAGCCCTTGCTGCATCCCCCACATAAGACGTTCTGCACTCTCTCCTTCCCAGCTGCCTCCCTGGGGATCTCTGTAATAGGTCAGCCCTTGACAGCTGATGCGGCTCTCTGGCCATTCTGTCTCCTCCAGGAACATCCGTGTAGCCATCCTGCCTTGCTCTGAAAGCAGCACCTGCTTGT ........................................................((((.(((....((((((.....((((((((.............................(((((((........)))))))...((((...(((.....)))..))))..)))))))).)))))))))..))))........................................................... ..................................................51..........................................................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TGACAGCTGATGCGGCTCTCTGGCCATT............................................................... | 28 | 1 | 9.00 | 9.00 | - | 1.00 | 3.00 | 4.00 | 1.00 | - | - | - | - |
| ...............................................................................................................................................................TGACAGCTGATGCGGCTCTCTGGCCAT................................................................ | 27 | 1 | 7.00 | 7.00 | - | 5.00 | 1.00 | - | 1.00 | - | - | - | - |
| .............................................TGGCAGGAAGAGCAGTTGGTACCCTAGA................................................................................................................................................................................. | 28 | 1 | 5.00 | 5.00 | 3.00 | - | - | - | - | 1.00 | 1.00 | - | - |
| .............................................TGGCAGGAAGAGCAGTTGGTACCCTAG.................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................ATCCTGCCTTGCTCTGAAAGC............. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - |
| ....................TAACAACAAAAAATTCAGAGAGCCATGGC......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................CTGGGGATCTCTGTAATAGGTCAGC.............................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGGCCATTCTGTCTCCTCCAGGAACATC........................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................TCCCCCACATAAGACGTTCTGCACTCTCTC....................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..................................TCAGAGAGCCATGGCAGGAAGAGCAGTT............................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TCCTGCCTTGCTCTGAAAGCAGCACCTGC.... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GTAGCCATCCTGCCTTGCTCTGAAAGC............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................TAAGACGTTCTGCACTCTCTCCTTCCC................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................TTGACAGCTGATGCGGCTCTCTGGCCAT................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................TGGGGATCTCTGTAATAGGTCAGC.............................................................................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| TAGGAAAAAAAGAAACAAAATAACAACAAAAAATTCAGAGAGCCATGGCAGGAAGAGCAGTTGGTACCCTAGAGCCCTTGCTGCATCCCCCACATAAGACGTTCTGCACTCTCTCCTTCCCAGCTGCCTCCCTGGGGATCTCTGTAATAGGTCAGCCCTTGACAGCTGATGCGGCTCTCTGGCCATTCTGTCTCCTCCAGGAACATCCGTGTAGCCATCCTGCCTTGCTCTGAAAGCAGCACCTGCTTGT ........................................................((((.(((....((((((.....((((((((.............................(((((((........)))))))...((((...(((.....)))..))))..)))))))).)))))))))..))))........................................................... ..................................................51..........................................................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................CCATTCTGTCTCCTCCA................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | 0.25 |