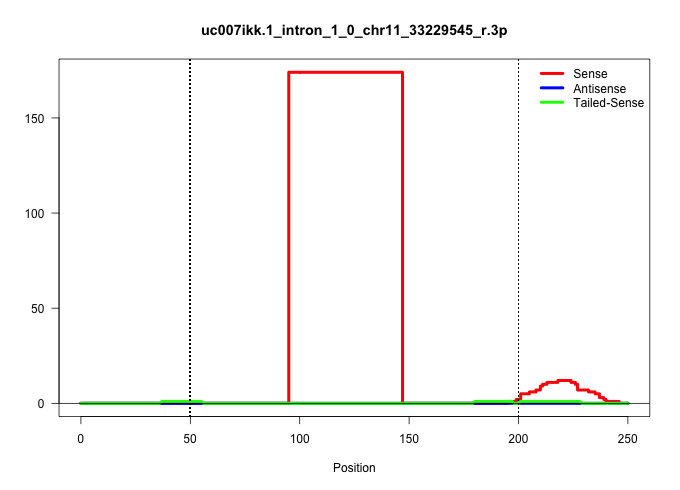
| Gene: Ranbp17 | ID: uc007ikk.1_intron_1_0_chr11_33229545_r.3p | SPECIES: mm9 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(12) TESTES |
| ATTCCAAAGTAGGAGTGTTTTCTGTGGGGAAAGGTACTGCAGTTGGGGATGACGTAGTTTTCATCTCATGAGCTCTGGACTCATTCCCTGTTTCAGTATGGATGTAGATCTTTGAGTTTTAGACTTGTTAACTTGAGCAGCCTTTTGAAACTACCTATGAATGATAAAGCTTTTAACACTTACATTTTTCATGCATGCAGGTGTATGCTCGTATGTCAGAAGTCTTAGGAATAACAGATGATAACCATGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................GTATGGATGTAGATCTTTGAGTTTTAGACTTGTTAACTTGAGCAGCCTTTTG....................................................................................................... | 52 | 1 | 174.00 | 174.00 | 174.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGTATGCTCGTATGTCAGAAGTCTTA....................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................GTATGTCAGAAGTCTTAGGAATAACAG............. | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGCTCGTATGTCAGAAGTCTTAGGAAT.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TATGTCAGAAGTCTTAGGAATAACAGAT........... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................TCGTATGTCAGAAGTCTTAGGAATAAC............... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................TGTATGCTCGTATGTCAGAAGTCTTAGt..................... | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGTGTATGCTCGTATGTCAGAAGTCTT........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GAAGTCTTAGGAATAACAGATGATAACC.... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TACATTTTTCATGCtgga.................................................... | 18 | tgga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGGTGTATGCTCGTATGTCAGAAGTC.......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGTCAGAAGTCTTAGGAATAACAGATG.......... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................TGCAGTTGGGGATGAggct.................................................................................................................................................................................................. | 19 | ggct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ATTCCAAAGTAGGAGTGTTTTCTGTGGGGAAAGGTACTGCAGTTGGGGATGACGTAGTTTTCATCTCATGAGCTCTGGACTCATTCCCTGTTTCAGTATGGATGTAGATCTTTGAGTTTTAGACTTGTTAACTTGAGCAGCCTTTTGAAACTACCTATGAATGATAAAGCTTTTAACACTTACATTTTTCATGCATGCAGGTGTATGCTCGTATGTCAGAAGTCTTAGGAATAACAGATGATAACCATGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................CAGTATGGATGTAGATtgtg............................................................................................................................................. | 20 | tgtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |