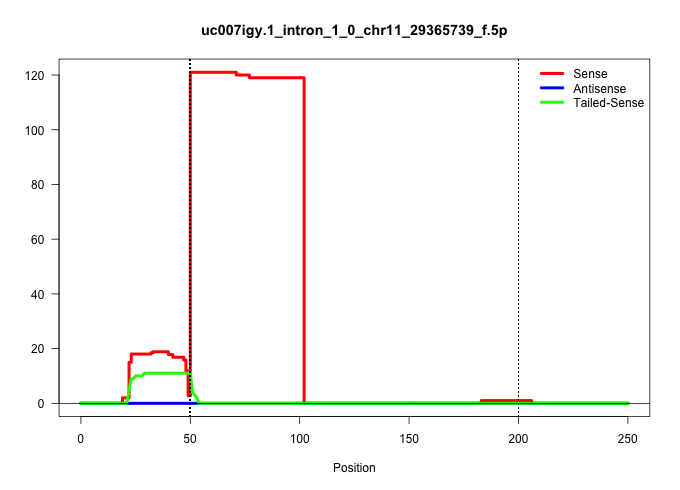
| Gene: Ccdc88a | ID: uc007igy.1_intron_1_0_chr11_29365739_f.5p | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(18) TESTES |
| GGTTTAAATAAAGAGCGGCTCCTACACGATGAGCAGAGCACTGATGACAGGTAAGTGGGATGTGGAGAAAGTTCTGAAAGATGTGGACTGAAAGGAAATTTAGCTTTATTAATGGTTGATTCAGACTTGTTTTACATTTCTGTTTTTGGTTAATTATAGTCTTTTAATTAGGATTAATGTGATTTTTAAGATCTACAGAGCCACTTGTTTCAGATAGACTAGAGATCTACTCTTTTGTGGGCCAGTAAGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGGGATGTGGAGAAAGTTCTGAAAGATGTGGACTGAAAGGAAATTTA.................................................................................................................................................... | 52 | 1 | 119.00 | 119.00 | 119.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACACGATGAGCAGAGCACTGATGACA......................................................................................................................................................................................................... | 27 | 1 | 9.00 | 9.00 | - | 2.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - |
| .......................ACACGATGAGCAGAGCACTGATGACAGt....................................................................................................................................................................................................... | 28 | t | 3.00 | 0.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACACGATGAGCAGAGCACTGATGAC.......................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................TACACGATGAGCAGAGCACTGATGACAGt....................................................................................................................................................................................................... | 29 | t | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................ACGATGAGCAGAGCACTGATGACAGt....................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................ACACGATGAGCAGAGCACTGATGAC.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................TCCTACACGATGAGCAGAGCACTGATGA........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACACGATGAGCAGAGCACTGATGACAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACACGATGAGCAGAGCACT................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACACGATGAGCAGAGCACTGATGACAGta...................................................................................................................................................................................................... | 30 | ta | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................TACACGATGAGCAGAGCACTGATGACAGtagt.................................................................................................................................................................................................... | 32 | tagt | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGGGATGTGGAGAAAG................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TTTTAAGATCTACAGAGCCACTT............................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................ACACGATGAGCAGAGCA.................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TCCTACACGATGAGCAGAGCACTGATG............................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................ACACGATGAGCAGAGCACTGATGACAGtat..................................................................................................................................................................................................... | 30 | tat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGAGCAGAGCACTGATGACAGtagg.................................................................................................................................................................................................... | 25 | tagg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................ACAGGTAAGTGGGATGTGGAGAAAGTTCTGA............................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................GCAGAGCACTGATGACAG........................................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .................................CAGAGCACTGATGACAG........................................................................................................................................................................................................ | 17 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGTTTAAATAAAGAGCGGCTCCTACACGATGAGCAGAGCACTGATGACAGGTAAGTGGGATGTGGAGAAAGTTCTGAAAGATGTGGACTGAAAGGAAATTTAGCTTTATTAATGGTTGATTCAGACTTGTTTTACATTTCTGTTTTTGGTTAATTATAGTCTTTTAATTAGGATTAATGTGATTTTTAAGATCTACAGAGCCACTTGTTTCAGATAGACTAGAGATCTACTCTTTTGTGGGCCAGTAAGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|