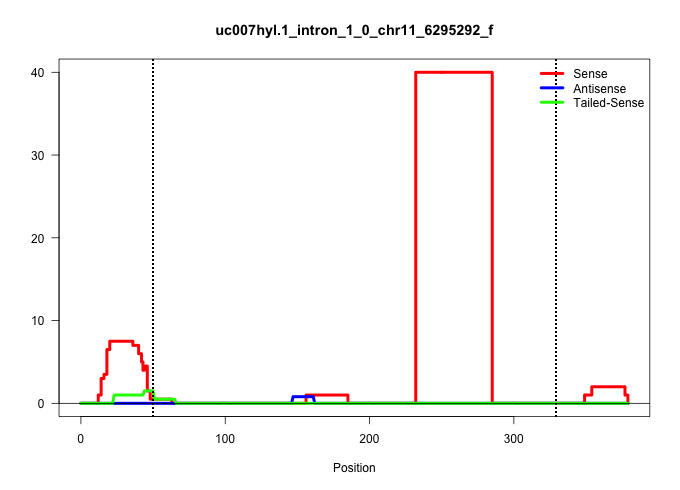
| Gene: Zmiz2 | ID: uc007hyl.1_intron_1_0_chr11_6295292_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
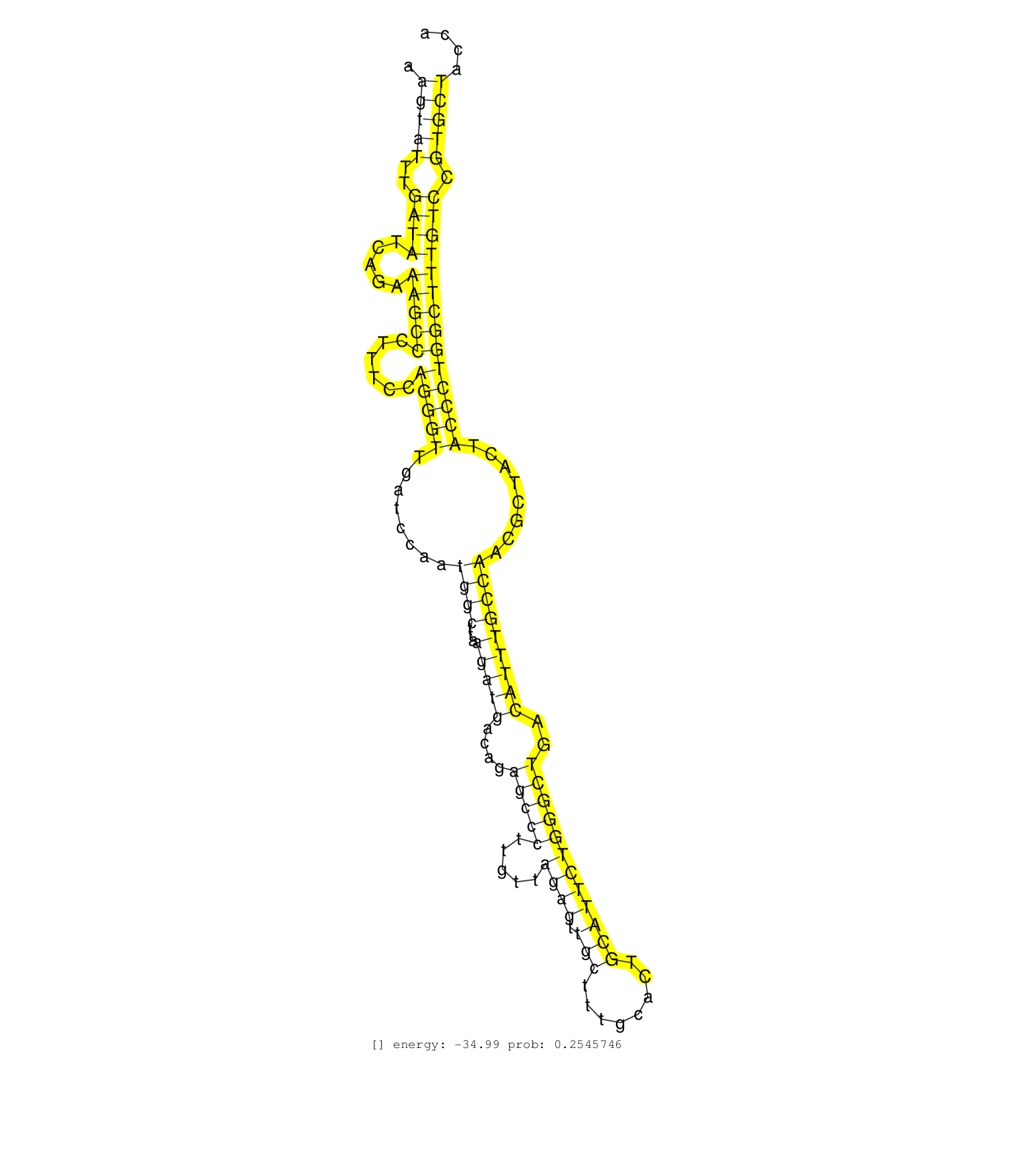
| GACTTTTCAGCATCTGCCTCAAAGAACACTCAAGCTCTGCTCCTTCTCAGGTCTCTGGGCTCCGCAGGGTGGGGGCTACCCCTCTGACCTTCTCCAGGGCCTGCTGAGGGTTCCTGGCTTCCTATGATTTATATGGGAAAGGGAATGTGGGAAGTATTTGATATCAGAAAGCCCTTTCCAGGGTTGATCCAATGGCTTAAGATGACAGAGCCCTTGTTAGAGTTGCTTTGCACTGCATTCTGGGCTGACATTTGCCAACGCTACTACCCTGGCTTTGTCCGTGCTACCATGAGCATCTAAGCTCCTGCCTTCTCCCTCCTGTCCCGCAGCCGGAGCAGCAACTCCCGGATAAGCTCCAGCACATTGGGCCTGTAGGCTG ........................................................................................................................................................(((((..((((.....(((((......(((((........((((...(((((....(((((.....((((.(((........))))))))))))..)))))))))........)))))))))))))).))))).............................................................................................. .......................................................................................................................................................152......................................................................................................................................289........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................CTGCATTCTGGGCTGACATTTGCCAACGCTACTACCCTGGCTTTGTCCGTGC............................................................................................... | 52 | 1 | 40.00 | 40.00 | 40.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................AACTCCCGGATAAGCTCCAGCACATTGG............ | 28 | 1 | 4.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TCAAAGAACACTCAAGCTCTGCTCCTTC............................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................TCAAAGAACACTCAAGCTCTGCTCCTTCTCAG......................................................................................................................................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............TCTGCCTCAAAGAACACTCAAGCTCTGCTCC................................................................................................................................................................................................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................TAAGCTCCAGCACATTGGGCCTGTAGGC.. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................GAACACTCAAGCTCTGCTCCTTCTCAGc........................................................................................................................................................................................................................................................................................................................................ | 28 | c | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGCCTCAAAGAACACTCAAGCTCTGC................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............TGCCTCAAAGAACACTCAAGCTCTGCTC................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............GCCTCAAAGAACACTCAAGC........................................................................................................................................................................................................................................................................................................................................................ | 20 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TTTGATATCAGAAAGCCCTTTCCAGGGTT.................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................TCCAGCACATTGGGCCTGTAGGCTG | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................AAAGAACACTCAAGCTCTGCTCCTTCTC........................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TCTCAGGTCTCTGGGCTCCtct......................................................................................................................................................................................................................................................................................................................... | 22 | tct | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ................CCTCAAAGAACACTCAAGCT....................................................................................................................................................................................................................................................................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ............................................TCTCAGGTCTCTGGGCTCC............................................................................................................................................................................................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| GACTTTTCAGCATCTGCCTCAAAGAACACTCAAGCTCTGCTCCTTCTCAGGTCTCTGGGCTCCGCAGGGTGGGGGCTACCCCTCTGACCTTCTCCAGGGCCTGCTGAGGGTTCCTGGCTTCCTATGATTTATATGGGAAAGGGAATGTGGGAAGTATTTGATATCAGAAAGCCCTTTCCAGGGTTGATCCAATGGCTTAAGATGACAGAGCCCTTGTTAGAGTTGCTTTGCACTGCATTCTGGGCTGACATTTGCCAACGCTACTACCCTGGCTTTGTCCGTGCTACCATGAGCATCTAAGCTCCTGCCTTCTCCCTCCTGTCCCGCAGCCGGAGCAGCAACTCCCGGATAAGCTCCAGCACATTGGGCCTGTAGGCTG ........................................................................................................................................................(((((..((((.....(((((......(((((........((((...(((((....(((((.....((((.(((........))))))))))))..)))))))))........)))))))))))))).))))).............................................................................................. .......................................................................................................................................................152......................................................................................................................................289........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................GCTCCTTCTCAGGTCTCTGGGCTCCGCA......................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................CCCGCAGCCGGAGCcgcg........................................... | 18 | cgcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |