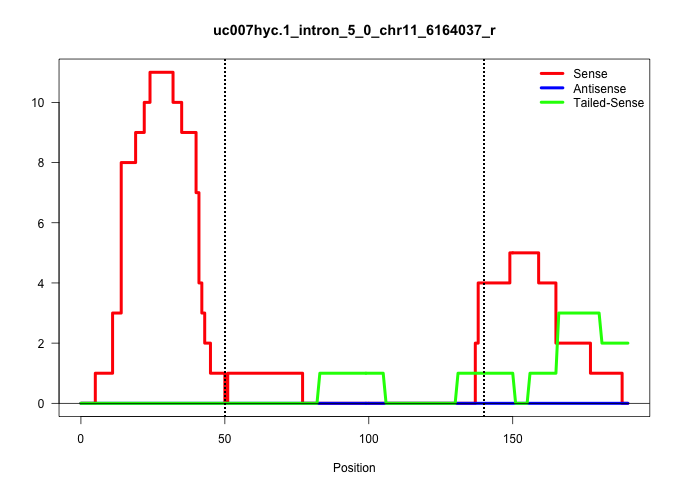
| Gene: Ddx56 | ID: uc007hyc.1_intron_5_0_chr11_6164037_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(18) TESTES |
| AATTTTGACCTTCCTCCCACCGCAGAGGCTTATGTCCATCGAGCTGGCAGGTAGGAGGATGGGGCGTGGGGTGAGCTGGGGCGTGCGGAAGGACAGATCCTGTGGGCTAGTGGTACACGCTTGATCTTCTCCCTCTGCAGGACAGCTCGAGCCAACAACCCAGGCATAGTTCTGACCTTTGTGTTGCCCG ..............................................................(((.........)))..(((.((((((........))).))).))).................................................................................. ..................................................51........................................................109............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................CAGGACAGCTCGAGCCAACAACCCAGGC......................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............TCCCACCGCAGAGGCTTATGTCCATCG..................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TAGTTCTGACCTTTGTGTTGCCCGt | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............TCCCACCGCAGAGGCTTATGTCCATCGAG................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GAGGCTTATGTCCATCGAGCTGGCAG............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGACAGCTCGAGCCAACAACCCAGGCA........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ACAGCTCGAGCCAACAACCCAGGCATAGTT................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................GTGGGCTAGTGGTACACGC...................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................TAGTTCTGACCTTTGTGTTGCCCGctga | 28 | ctga | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGACAGCTCGAGCCAACAAC............................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TAGTTCTGACCTTTGTGTTGCC.. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACCTTCCTCCCACCGCAGAGGCTTATGT........................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGATCTTCTCCCTCTGCAGagt............................................... | 22 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TAGGAGGATGGGGCGTGGGGTGAGCT................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGCGGAAGGACAGATCCTGTGtt.................................................................................... | 23 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............TCCCACCGCAGAGGCTTATGTCCATC...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CAGAGGCTTATGTCCATCG..................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CCGCAGAGGCTTATGTCCATCGAGCT................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........TCCTCCCACCGCAGAGGCTTA.............................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............TCCCACCGCAGAGGCTTATGTCCATCGA.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AGCCAACAACCCAGGCATAGTTCTGACC............. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CCTCTGCAGGACAGCTggcc....................................... | 20 | ggcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........TCCTCCCACCGCAGAGGCTTATGTCCATC...................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................AACCCAGGCATAGTTCTGACCTTTt......... | 25 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AATTTTGACCTTCCTCCCACCGCAGAGGCTTATGTCCATCGAGCTGGCAGGTAGGAGGATGGGGCGTGGGGTGAGCTGGGGCGTGCGGAAGGACAGATCCTGTGGGCTAGTGGTACACGCTTGATCTTCTCCCTCTGCAGGACAGCTCGAGCCAACAACCCAGGCATAGTTCTGACCTTTGTGTTGCCCG ..............................................................(((.........)))..(((.((((((........))).))).))).................................................................................. ..................................................51........................................................109............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................CCCAGGCATAGTTCTGAcag............... | 20 | cag | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CCCAGGCATAGTTCTcagg................. | 19 | cagg | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................ACCCAGGCATAGTTCagg.................. | 18 | agg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CCCAGGCATAGTTcagt................... | 17 | cagt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |