
| Gene: Nf2 | ID: uc007hvj.1_intron_1_0_chr11_4716169_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(11) TESTES |
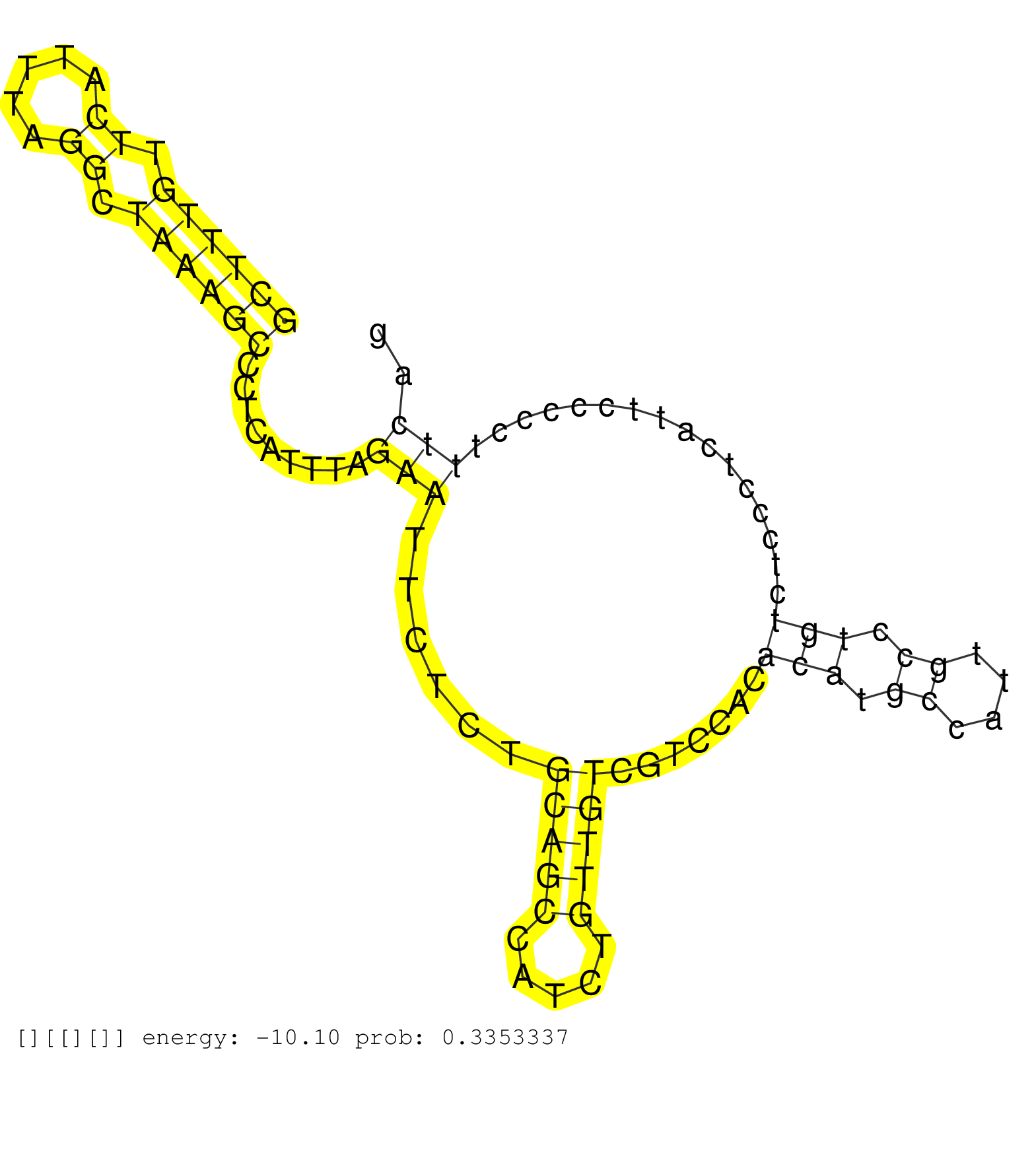
| ACGCAGCTGTCTTACCGATAGCTTCTTAGTGGCTGAACTAGCAACTGGTCTGCAACAGTTTTTTAAAAAGTTCCCTTTGTCGGAAACTCACTAACAGGCTGCTTTGTTCATTTAGGCTAAAGCCCTCATTTAGAATTCTCTGCAGCCATCTGTTGTCGTCCACACATGCCATTGCCTGTCTCCCTCATTCCCCCTTTCAGGTGAAGAAGCAGATTTTGGATGAAAAGGTCTACTGCCCTCCCGAGGCGTC ....................................................................................................((((((.((.....)).)))))).........(((......(((((.....))))).......(((.((....)).)))................))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................CTTTGTCGGAAACTCACTAACAGGCTGCTTTGTTCATTTAGGCTAAAGCCCT............................................................................................................................ | 52 | 1 | 50.00 | 50.00 | 50.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GAAGCAGATTTTGGATGAAA......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................TGGATGAAAAGGTCTACTGCCCTCCC........ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................ATTTAGAATTCTCgc............................................................................................................ | 15 | gc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................TCGGAAACTCACTAACAGGC....................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GATTTTGGATGAAAAGGTCTACcccc............. | 26 | cccc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................TGAAAAGGTCTACTGCCCTCCCGAGGCGTC | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGATTTTGGATGAAAAGGTCTACTGCCCTC.......... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGAAAAGGTCTACTGCCCTCCCGAGGCGT. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGCAGATTTTGGATGAAAAGGTCTAC................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ACGCAGCTGTCTTACCGATAGCTTCTTAGTGGCTGAACTAGCAACTGGTCTGCAACAGTTTTTTAAAAAGTTCCCTTTGTCGGAAACTCACTAACAGGCTGCTTTGTTCATTTAGGCTAAAGCCCTCATTTAGAATTCTCTGCAGCCATCTGTTGTCGTCCACACATGCCATTGCCTGTCTCCCTCATTCCCCCTTTCAGGTGAAGAAGCAGATTTTGGATGAAAAGGTCTACTGCCCTCCCGAGGCGTC ....................................................................................................((((((.((.....)).)))))).........(((......(((((.....))))).......(((.((....)).)))................))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TCTGCAGCCATCTGTaaca................................................................................................. | 19 | aaca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................TCTGCAGCCATCTGttta.................................................................................................. | 18 | ttta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |