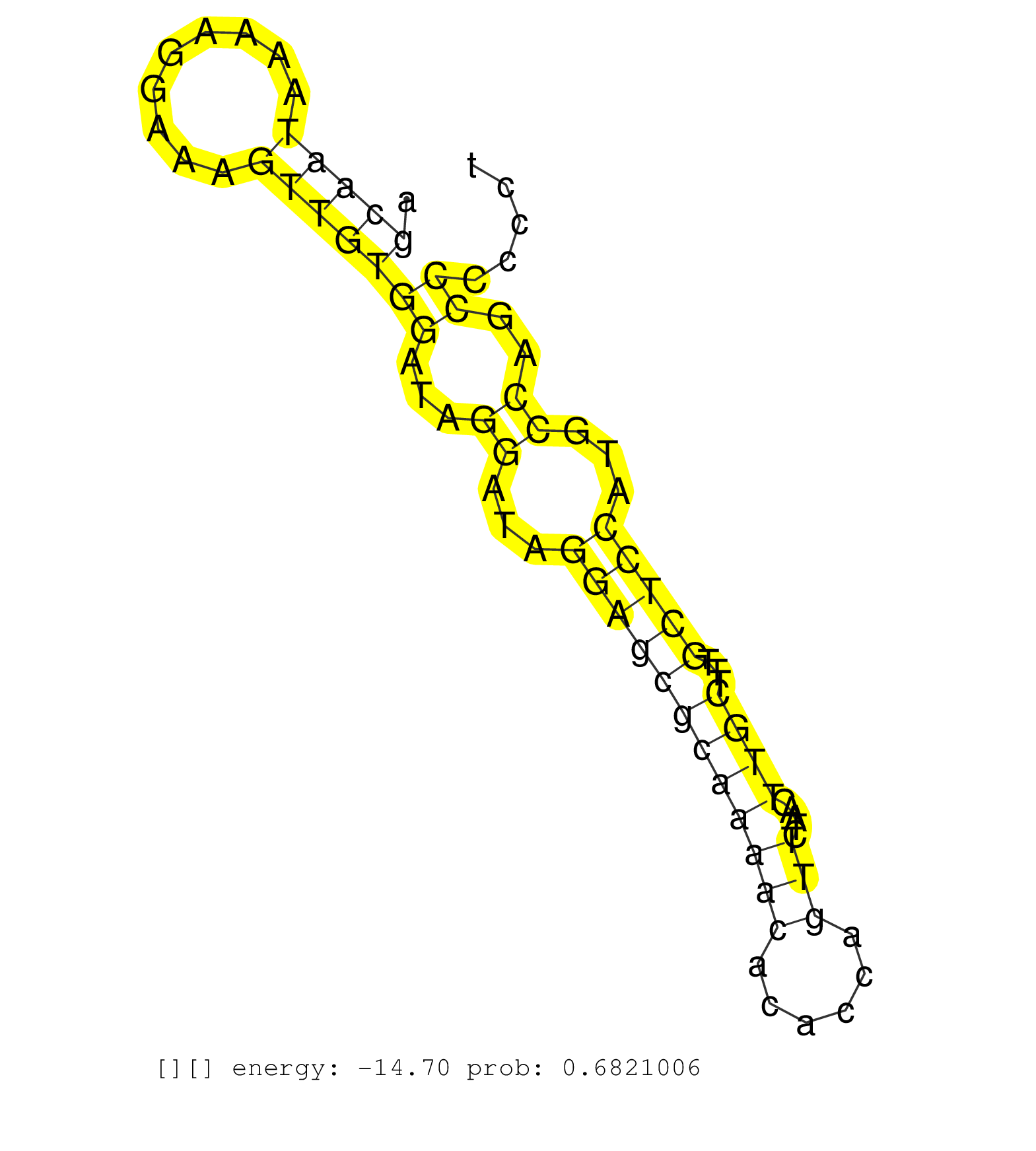

| Gene: 4921536K21Rik | ID: uc007htt.1_intron_1_0_chr11_3787057_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(22) TESTES |
| CCACCCTGAACCATGGCAGAATGATCTGCCCCAAGAACAGAACCACCCTGGTGAGAGCGGAGGAGCCCCAGAGCAGTTGGTCTCCACCATGGTAGAGCAATAAAAGGAAAGTTGTGGATAGGATAGGAGCGCAAAACACACCAGTTCTAACTTGCTTTGCTCCATGCCAGCCCCCCTATCCCTGTGAAATCCCCACAGCCACCTATGCTTGGTGGAAGAGCCAGTCTCCTTGGCTTGTGTAAACATGGAA ................................................................................................(((((.........)))))((...((...((((((((((((......))).....))))...)))))...))..)).............................................................................. ...............................................................................................96...............................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................TAAAAGGAAAGTTGTGGATAGGATAGGA.......................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....CCTGAACCATGGCAGAATGATC................................................................................................................................................................................................................................ | 22 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAAAAGGAAAGTTGTGGATAGGATAGGt.......................................................................................................................... | 28 | t | 3.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................TAAAAGGAAAGTTGTGGATAGGATAGGAGC........................................................................................................................ | 30 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| .........................................................................................TGGTAGAGCAATAAAAGGAAAGTTGTGGAT................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGGATAGGATAGGAGCGCAAAACACACC............................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................AACAGAACCACCCTGGTGAGAGCGGAt............................................................................................................................................................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TAGGATAGGAGCGCAAAACACACCAGT......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AAAAGGAAAGTTGTGGATAGGATAGGAG......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................TAGAGCAATAAAAGGAAAGTTGTGGA.................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAAAAGGAAAGTTGTGGATAGGATAGGc.......................................................................................................................... | 28 | c | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AGGAGCGCAAAACACACCAG.......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................AATAAAAGGAAAGTTGTGGATAGGATAGG........................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAAAAGGAAAGTTGTGGATAGGATAGGtt......................................................................................................................... | 29 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................ATAAAAGGAAAGTTGTGGATAGGATAGGAtt........................................................................................................................ | 31 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................TAAAAGGAAAGTTGTGGATAGGAT.............................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................ATAAAAGGAAAGTTGTGGATAGGAT.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TTCTAACTTGCTTTGCTCCATGCCAGCCC............................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGGTAGAGCAATAAAAGGAAAGTTGTG...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................CACCATGGTAGAGCA....................................................................................................................................................... | 15 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 |
| CCACCCTGAACCATGGCAGAATGATCTGCCCCAAGAACAGAACCACCCTGGTGAGAGCGGAGGAGCCCCAGAGCAGTTGGTCTCCACCATGGTAGAGCAATAAAAGGAAAGTTGTGGATAGGATAGGAGCGCAAAACACACCAGTTCTAACTTGCTTTGCTCCATGCCAGCCCCCCTATCCCTGTGAAATCCCCACAGCCACCTATGCTTGGTGGAAGAGCCAGTCTCCTTGGCTTGTGTAAACATGGAA ................................................................................................(((((.........)))))((...((...((((((((((((......))).....))))...)))))...))..)).............................................................................. ...............................................................................................96...............................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TGGAAGAGCCAGTCTggt....................... | 18 | ggt | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................ACAGAACCACCCTGctat........................................................................................................................................................................................................ | 18 | ctat | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................AAATCCCCACAGCCgttc.................................................. | 18 | gttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................AAATCCCCACAGCCAttgc................................................. | 19 | ttgc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |