
| Gene: Eif4enif1 | ID: uc007hsf.1_intron_11_0_chr11_3135664_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(12) TESTES |
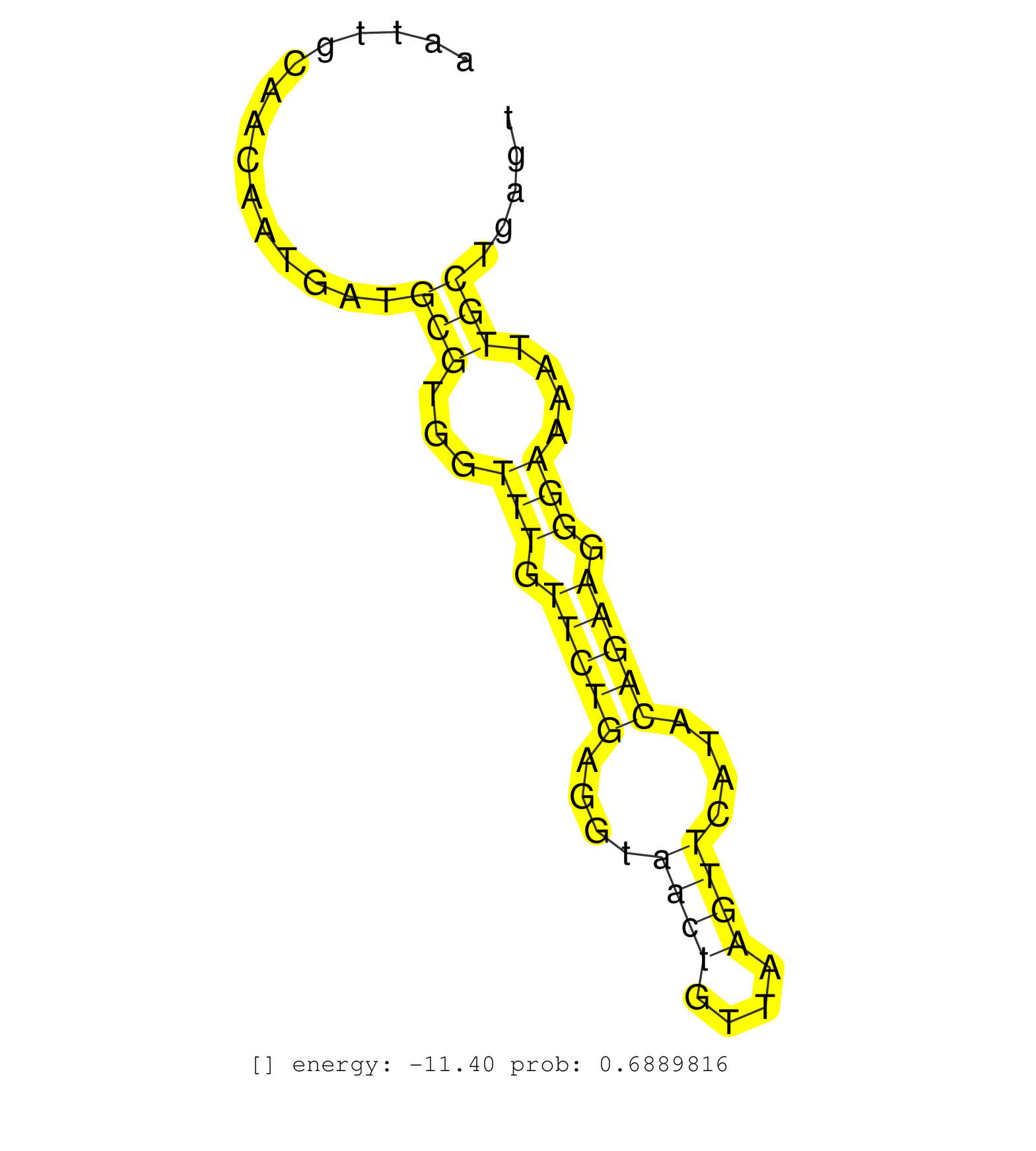
| GAGATCCATTCCAAGGCATGCGCAAGCCCATGAGTCCTGTCTCAGCCCAGGTCAGGCTGAGCTCTTTCTCTTTGCCTTTTCACTCTTCTCTTGCTTGTAGGGTAGAGAATTGAATTGCAACAATGATGCGTGGTTTGTTCTGAGGTAACTGTTAAGTTCATACAGAAGGGAAAATTGCTGAGTGATTAGTATTAAATTAAAATTTTTCTTCTTTAGATTTACTTATTTTCTGTGTATGAGTGTTTGCTTC ...............................................................................................................................(((...(((.(((((....((((....))))....))))).)))....)))........................................................................ ................................................................................................................113...................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................CAACAATGATGCGTGGTTTGTTCTGAGG......................................................................................................... | 28 | 1 | 33.00 | 33.00 | 8.00 | 7.00 | 5.00 | 4.00 | - | 3.00 | 2.00 | 2.00 | - | 2.00 | - | - |
| ......................................................................................................................AACAATGATGCGTGGTTTGTTCTGAGG......................................................................................................... | 27 | 1 | 8.00 | 8.00 | 4.00 | - | 3.00 | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................CAACAATGATGCGTGGTTTGTTCTGAGGT........................................................................................................ | 29 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAACAATGATGCGTGGTTTGTTCTGAGGTA....................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................................ATTTTCTGTGTATGAGTcctg..... | 21 | cctg | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - |
| ......................................................................................................................AACAATGATGCGTGGTTTGTTCTGAGGTAA...................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................CAACAATGATGCGTGGTTTGTTCTGcgg......................................................................................................... | 28 | cgg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................AACAATGATGCGTGGTTTGTTCTGAtg......................................................................................................... | 27 | tg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGCGTGGTTTGTTCatga.......................................................................................................... | 18 | atga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................AACAATGATGCGTGGTTTGTTCT............................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...ATCCATTCCAAGGCATGC..................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................GTTAAGTTCATACAGAAGGGAAAATTGCT....................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................AACAATGATGCGTGGTTTGTTCTGAGGT........................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAACAATGATGCGTGGTTTGTTCTGAGGTAA...................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................GGAAAATTGCTGAGT................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | 0.11 | - | - | - | - | - | - | - | - | - |
| GAGATCCATTCCAAGGCATGCGCAAGCCCATGAGTCCTGTCTCAGCCCAGGTCAGGCTGAGCTCTTTCTCTTTGCCTTTTCACTCTTCTCTTGCTTGTAGGGTAGAGAATTGAATTGCAACAATGATGCGTGGTTTGTTCTGAGGTAACTGTTAAGTTCATACAGAAGGGAAAATTGCTGAGTGATTAGTATTAAATTAAAATTTTTCTTCTTTAGATTTACTTATTTTCTGTGTATGAGTGTTTGCTTC ...............................................................................................................................(((...(((.(((((....((((....))))....))))).)))....)))........................................................................ ................................................................................................................113...................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|