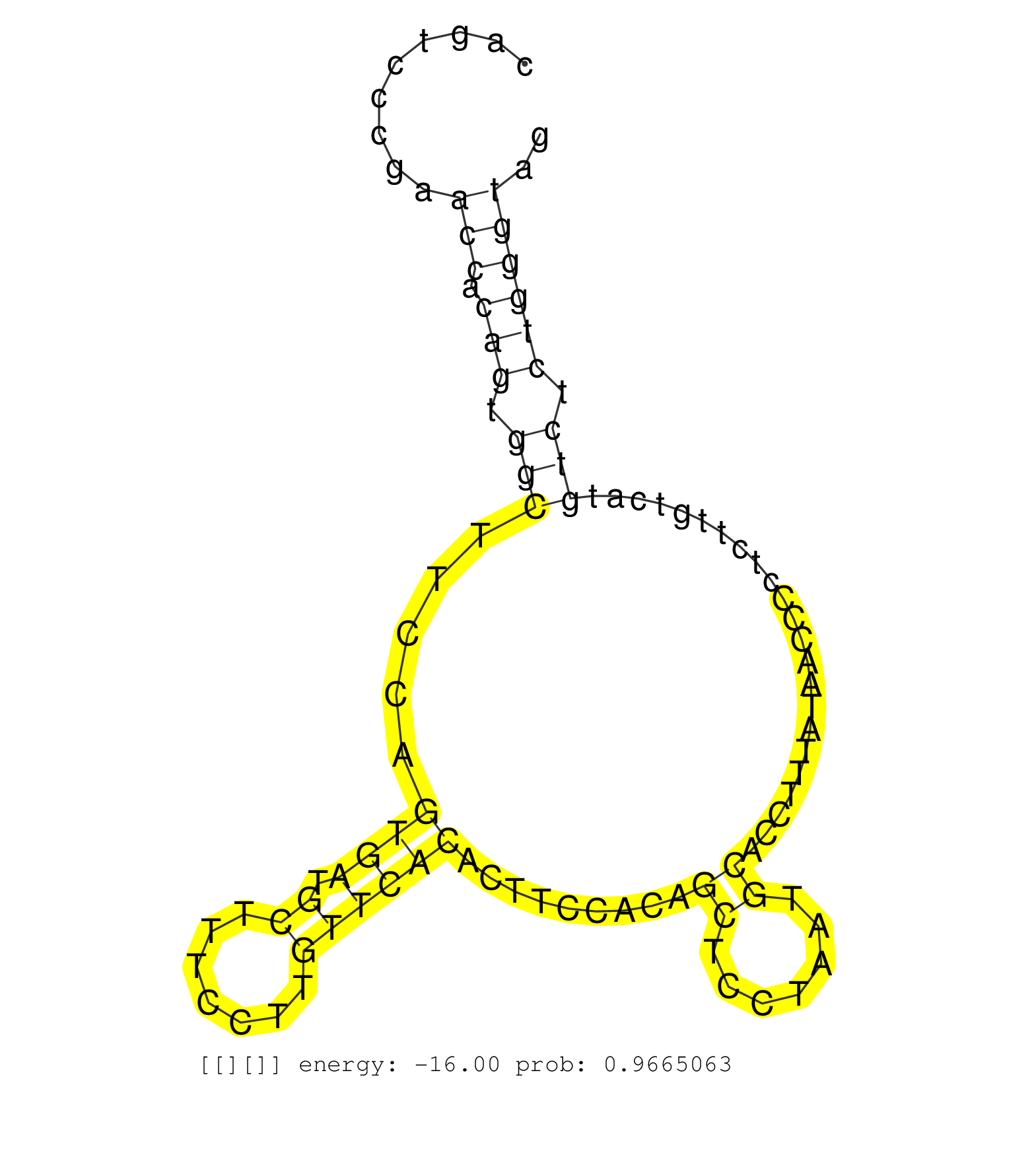

| Gene: Sfi1 | ID: uc007hrs.1_intron_20_0_chr11_3056868_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(16) TESTES |
| AGGCAACGCTGTGTTTTTTGTGTGAAGGCAGTGTTACTGAGCCTGCTGGGTAAGCTAGTTTCTATTAGGAACTTGGTAGCAAAACTGCAGAGAACTGGAGCAGTCCCGAACCACAGTGGCTTCCAGTGATGCTTTCCTTGTTCACACTTCCACAGCTCCTAATGCACCTTTATAACCCCTCTTGTCATGTCTCTGGGTAGTATTCTTGCTTTAGAGCCTTCAAAGACAACGTGACCCAGGCCCGACTACA .............................................................................................................(((.(((.(((.....((((.((.......)))))).........((.......)).......................))).)))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................CTTCCAGTGATGCTTTCCTTGTTCACACTTCCACAGCTCCTAATGCACCTTT............................................................................... | 52 | 1 | 115.00 | 115.00 | 115.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TAGGAACTTGGTAGCAAAACTGCAGA............................................................................................................................................................... | 26 | 1 | 10.00 | 10.00 | - | 5.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TAGGAACTTGGTAGCAAAACTGCAG................................................................................................................................................................ | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................TAGGAACTTGGTAGCAAAACTGCAGAG.............................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TAGAGCCTTCAAAGACAACGTGACCCA............ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................TATTAGGAACTTGGTAGCAAAACTGCAG................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TTAGGAACTTGGTAGCAAAACTGCAG................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................TGTTACTGAGCCTGCTGGGTAAGCTAGTT.............................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................TGTGTGAAGGCAGTGTTACTGAGCCTt............................................................................................................................................................................................................. | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................TATTAGGAACTTGGTAGCAAAACTGCAGA............................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TAGAGCCTTCAAAGACAACGTGACC.............. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................TATTAGGAACTTGGTAGCAAAACTGCAGAG.............................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................TATTAGGAACTTGGTAGCAAAACTGCA................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TATTAGGAACTTGGTAGCAAAACTGC.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TAGGAACTTGGTAGCAAAACTGCAGAGAAC........................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TTAGGAACTTGGTAGCAAAACTGCAGAGA............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................TAGAGCCTTCAAAGACAACGTGACCC............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGCAACGCTGTGTTTTTTGTGTGAAGGCAGTGTTACTGAGCCTGCTGGGTAAGCTAGTTTCTATTAGGAACTTGGTAGCAAAACTGCAGAGAACTGGAGCAGTCCCGAACCACAGTGGCTTCCAGTGATGCTTTCCTTGTTCACACTTCCACAGCTCCTAATGCACCTTTATAACCCCTCTTGTCATGTCTCTGGGTAGTATTCTTGCTTTAGAGCCTTCAAAGACAACGTGACCCAGGCCCGACTACA .............................................................................................................(((.(((.(((.....((((.((.......)))))).........((.......)).......................))).)))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................GAACTTGGTAGCAAAAttc...................................................................................................................................................................... | 19 | ttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................GGCTTCCAGTGATGCTTTCCTTGTTCA.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GCTTCCAGTGATGCTTTCCTTGTTCA.......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |