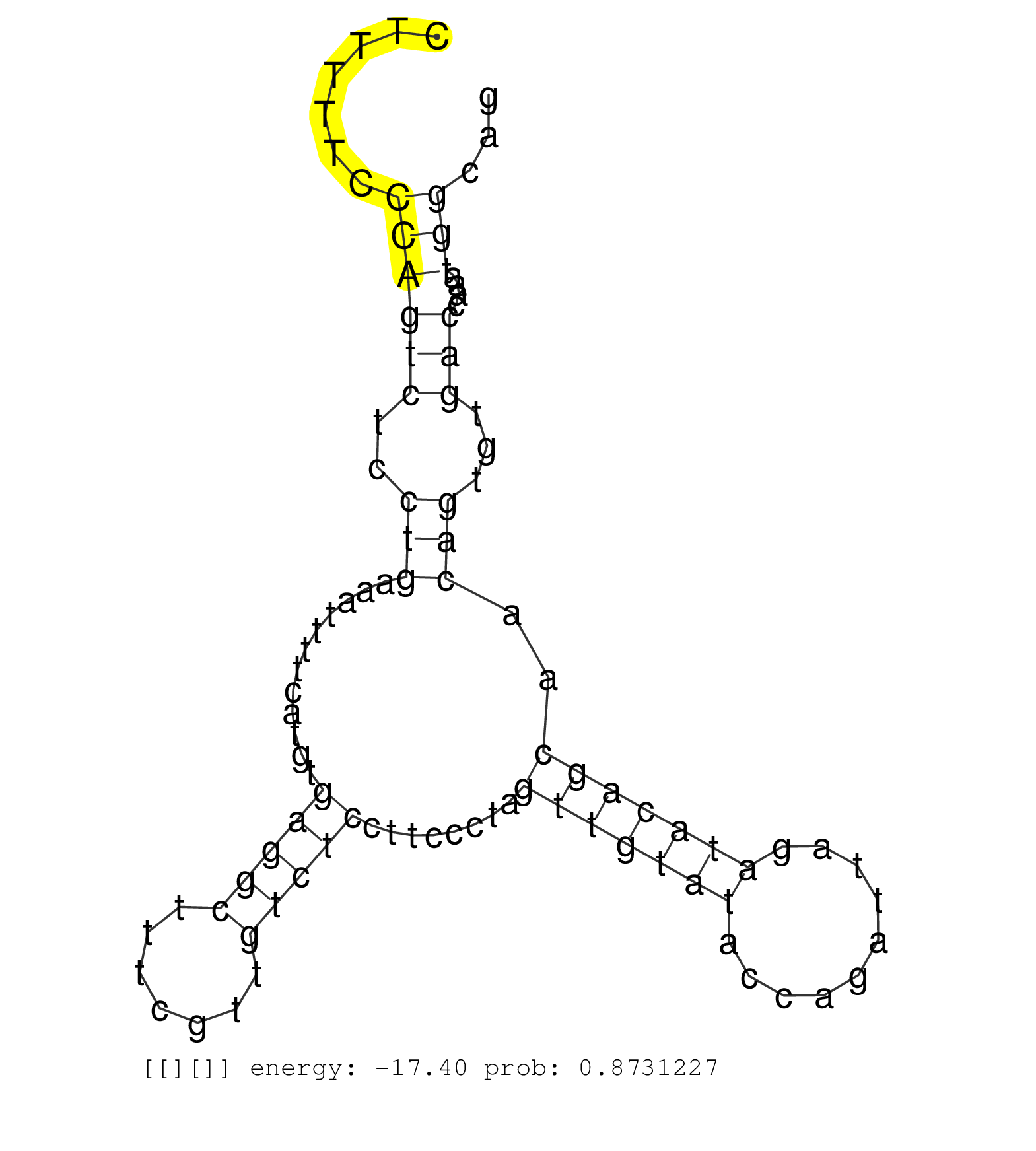

| Gene: Rab5b | ID: uc007hnv.1_intron_1_0_chr10_128118536_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(8) TESTES |
| TGGCAGGGAACAAAGCTGACCTTGCCAACAAGCGCATGGTGGAGTATGAAGTAAGGTGGCCTACAGAATCCCTAGCACACTTGGTCTGTATGTGGGACCTCTTTTTCCCAGTCTCCTGAAATTTTCATGTGAGGCTTTCGTTGTCTCCTTCCCTAGTTGTATACCAGATTAGATACAGCAACAGTGTGACCAAATGGCAGCCAACAGATTTTAAAGGAATCAGAAGAAAGTTCCAGATTAGAGGGGTAGG ...........................................................................................................((((((..(((............(((((.......)))))........(((((((..........)))))))..)))...)))....)))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................ACAGAATCCCTAGCACACTTGGTCTGTATGTGGGACCTCTTTTTCCCAGTCT........................................................................................................................................ | 52 | 1 | 32.00 | 32.00 | 32.00 | - | - | - | - | - | - | - |
| ...........................ACAAGCGCATGGTGGAGTATGAAGagt.................................................................................................................................................................................................... | 27 | agt | 4.00 | 0.00 | - | 1.00 | - | 2.00 | 1.00 | - | - | - |
| ...........................ACAAGCGCATGGTGGAGTATGAAGaa..................................................................................................................................................................................................... | 26 | aa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |
| ...........AAAGCTGACCTTGCCAACAAGCGCATGGTGt................................................................................................................................................................................................................ | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |
| ...........................ACAAGCGCATGGTGGAGTATGAAGaggt................................................................................................................................................................................................... | 28 | aggt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |
| ......................TGCCAACAAGCGCATGGTGGAGTATGAAGT...................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ......................TGCCAACAAGCGCATGGTGGAGTAT........................................................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 |
| ......................TGCCAACAAGCGCATGGTGGAGTATGAA........................................................................................................................................................................................................ | 28 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - |
| TGGCAGGGAACAAAGCTGACCTTGCCAACAAGCGCATGGTGGAGTATGAAGTAAGGTGGCCTACAGAATCCCTAGCACACTTGGTCTGTATGTGGGACCTCTTTTTCCCAGTCTCCTGAAATTTTCATGTGAGGCTTTCGTTGTCTCCTTCCCTAGTTGTATACCAGATTAGATACAGCAACAGTGTGACCAAATGGCAGCCAACAGATTTTAAAGGAATCAGAAGAAAGTTCCAGATTAGAGGGGTAGG ...........................................................................................................((((((..(((............(((((.......)))))........(((((((..........)))))))..)))...)))....)))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAGGGAACAAAGCTGactt........................................................................................................................................................................................................................................ | 19 | actt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - |
| ...........CTGACCTTGCCAACgtat............................................................................................................................................................................................................................. | 18 | gtat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - |