
| Gene: Mbc2 | ID: uc007hng.1_intron_4_0_chr10_127949024_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(1) PIWI.mut |
(15) TESTES |
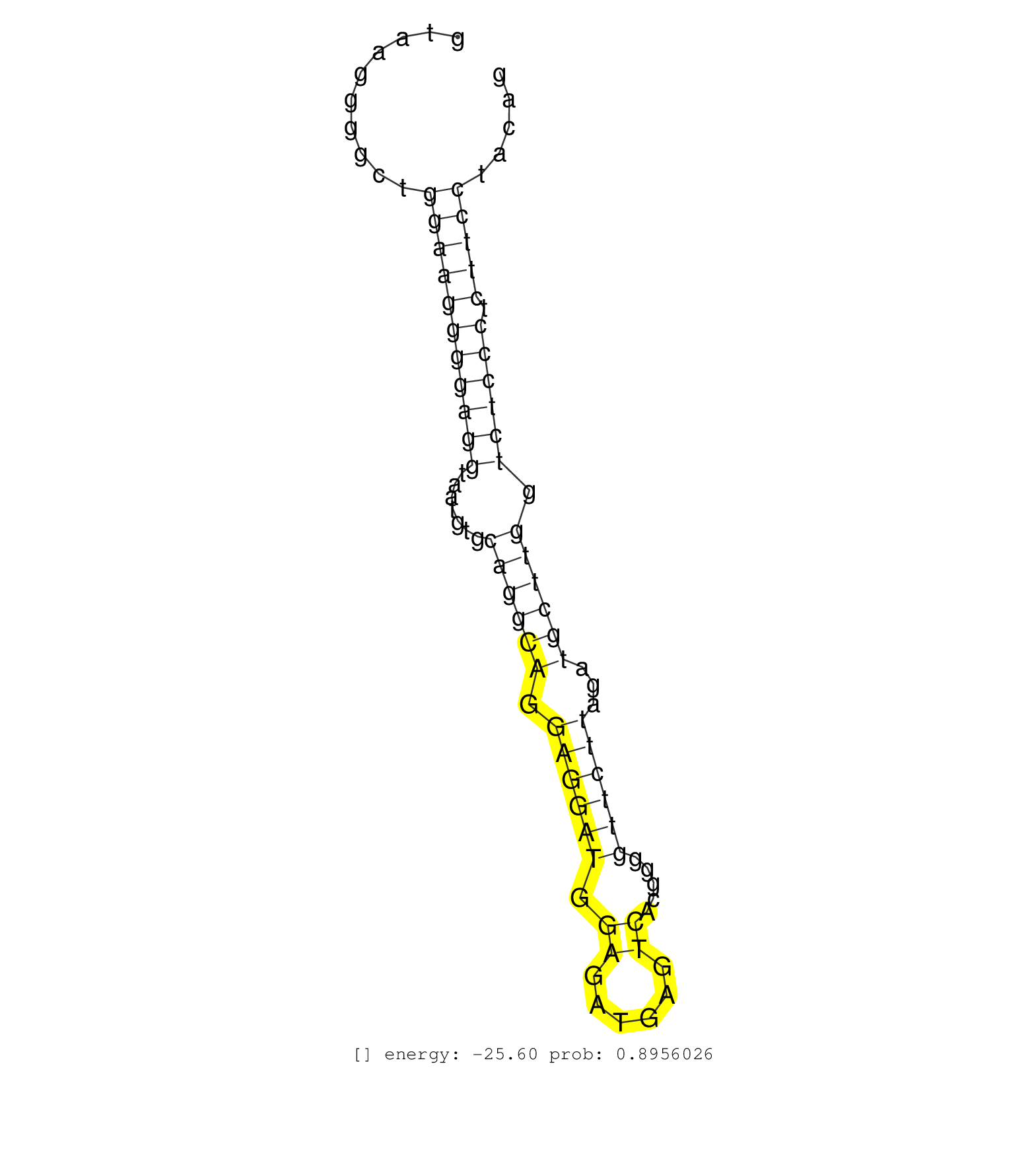
| CCTGCCTCCCCGGTCCTAGAGGTCAGGCATCGCCTAACACATGGTGACAGGTAAGGGGCTGGAAGGGGAGGTAATGTGCAGGCAGGAGGATGGAGATGAGTCACGGGGTTCTTAGATGCTTGGTCTCCCTCTTCCTACAGTCCCTCTGAGGCTCCCGTTGGGCCCCTGGGCCAGGTGAAGCTGACAGTGT ............................................................(((((((((((.......((((((.((((((.((......)).....))))))...)))))).)))))).)))))....................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037903(GSM510439) testes_rep4. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR037900(GSM510436) testes_rep1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................CAGGAGGATGGAGATGAGTCA....................................................................................... | 21 | 1 | 8.00 | 8.00 | 2.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 |
| ....................................................................................................................................TCCTACAGTCCCTCgatc........................................ | 18 | gatc | 7.00 | 0.00 | - | 2.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCctgc........................................ | 18 | ctgc | 3.00 | 0.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCgttc........................................ | 18 | gttc | 2.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................CAGGAGGATGGAGATGAGTCAt...................................................................................... | 22 | t | 2.00 | 8.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCgctc........................................ | 18 | gctc | 2.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCgagc........................................ | 18 | gagc | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................CAGGAGGATGGAGATGAGTt........................................................................................ | 20 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCcttc........................................ | 18 | cttc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TCCTAGAGGTCAGGCATCGCCTAACACAT.................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................GCAGGAGGATGGAGATGAGTCA....................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................CAGGAGGATGGAGATGAG.......................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCcgta........................................ | 18 | cgta | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................AGGCAGGAGGATGGAGATGAGTC........................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GGCAGGAGGATGGAGATGAG.......................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCCTACAGTCCCTCgcgc........................................ | 18 | gcgc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................CAGGAGGATGGAGATGAGTC........................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................GCAGGAGGATGGAGATGtg.......................................................................................... | 19 | tg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................CAGGAGGATGGAGATGAGTCAa...................................................................................... | 22 | a | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GCAGGAGGATGGAGATGAGTta....................................................................................... | 22 | ta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GGCAGGAGGATGGAGATGAGTC........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CCTGCCTCCCCGGTCCTAGAGGTCAGGCATCGCCTAACACATGGTGACAGGTAAGGGGCTGGAAGGGGAGGTAATGTGCAGGCAGGAGGATGGAGATGAGTCACGGGGTTCTTAGATGCTTGGTCTCCCTCTTCCTACAGTCCCTCTGAGGCTCCCGTTGGGCCCCTGGGCCAGGTGAAGCTGACAGTGT ............................................................(((((((((((.......((((((.((((((.((......)).....))))))...)))))).)))))).)))))....................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037903(GSM510439) testes_rep4. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR037900(GSM510436) testes_rep1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................ATGCTTGGTCTCCCTCTTCCTACAGTCCCTCTG.......................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................GCTTGGTCTCCCTCTTCCTACAGTCC............................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................GCTTGGTCTCCCgggg............................................................. | 16 | gggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................TACAGTCCCTCTGAGGC...................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |