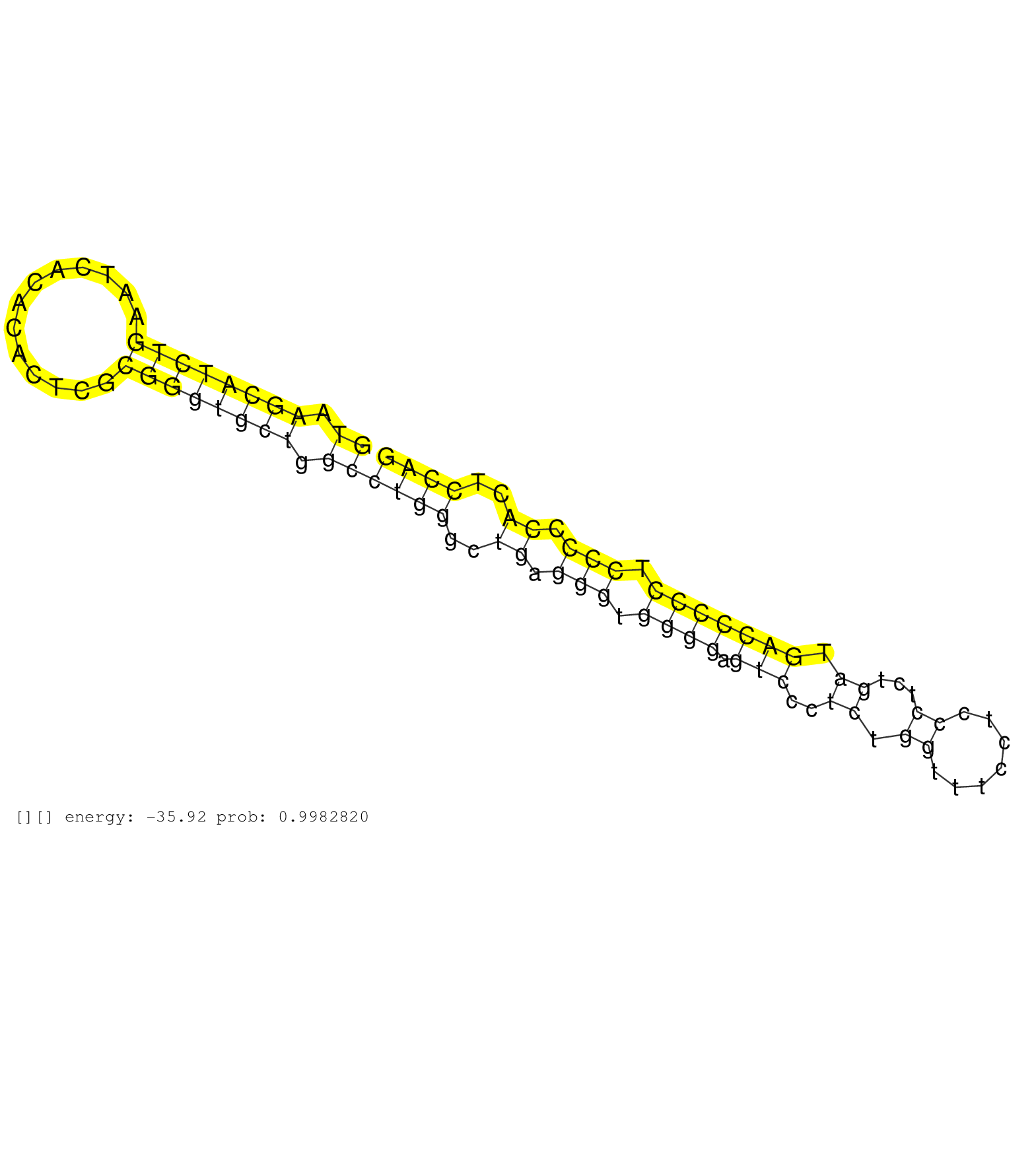

| Gene: Ankrd52 | ID: uc007hmm.1_intron_21_0_chr10_127826045_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| CCGTTCTCATACTTGGAGGGAAACCCCTTCACTCCTTTGCACTGTGCAGTGTAAGCATCTGAATCACACACTCGCGGGTGCTGGCCTGGGCTGAGGGTGGGGAGTCCCTCTGGTTTCCTCCCTCTGATGACCCCCTCCCCCACTCCAGAATTAATAACCAGGACAGCACCACAGAGATGCTGCTGGGGGCTCTGGGTG ..................................................((.((((((((.............)))))))).))((((..((.(((.((((.(((..((.((.......))...)).))))))).))).))..)))).................................................. ..................................................51...............................................................................................148................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGCATCTGAATCACACACTCGCGG......................................................................................................................... | 27 | 1 | 12.00 | 12.00 | 2.00 | 2.00 | 1.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - |
| ..................................................GTAAGCATCTGAATCACACACTCGCGGG........................................................................................................................ | 28 | 1 | 6.00 | 6.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGCATCTGAATCACACACTCGCG.......................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TTAATAACCAGGACAGCACCACAGAGA..................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAATAACCAGGACAGCACCACAGAGA..................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TAATAACCAGGACAGCACCACAGAGATG................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................TGACCCCCTCCCCCACTCCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGCATCTGAATCACACACTCGCGGGT....................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCATCTGAATCACACACTCGC........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................CATCTGAATCACACACTCGCGGGTGCTGGC................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GCTGCTGGGGGCTCTGtat. | 19 | tat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TTAATAACCAGGACAGCACCACAGAG...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGCATCTGAATCACACACTCGCGG......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGGGCTGAGGGTGGGGAGTCCCTC........................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GCCTGGGCTGAGGGTGGGGAttc............................................................................................ | 23 | ttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| CCGTTCTCATACTTGGAGGGAAACCCCTTCACTCCTTTGCACTGTGCAGTGTAAGCATCTGAATCACACACTCGCGGGTGCTGGCCTGGGCTGAGGGTGGGGAGTCCCTCTGGTTTCCTCCCTCTGATGACCCCCTCCCCCACTCCAGAATTAATAACCAGGACAGCACCACAGAGATGCTGCTGGGGGCTCTGGGTG ..................................................((.((((((((.............)))))))).))((((..((.(((.((((.(((..((.((.......))...)).))))))).))).))..)))).................................................. ..................................................51...............................................................................................148................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CCCCCACTCCAGAATTAATAACCA...................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |