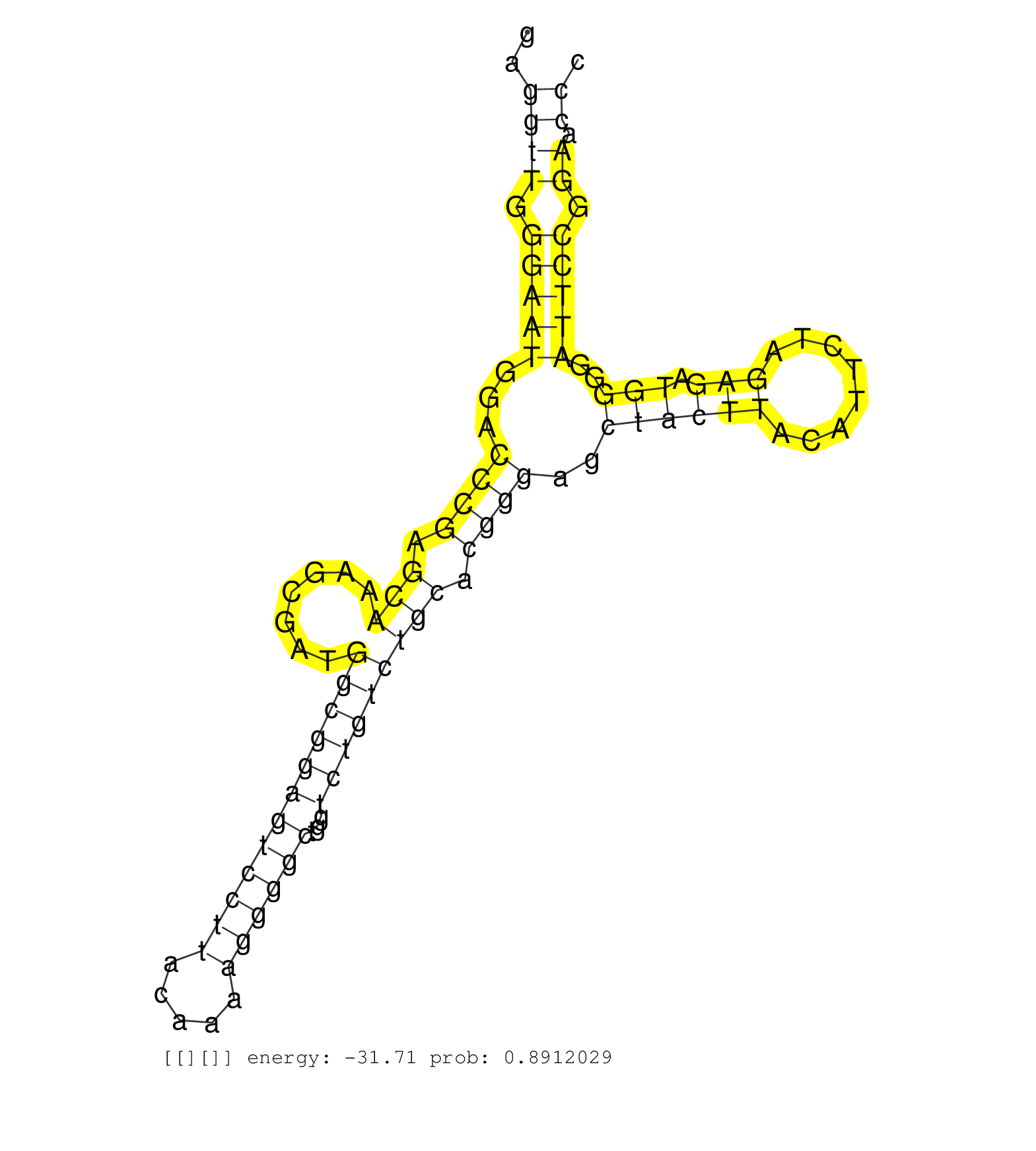

| Gene: Tmem4 | ID: uc007hmi.1_intron_0_0_chr10_127759832_f.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(22) TESTES |
| TCCGGCTGCTTTCCAGAGCCTGGGCCACGGCGGCGGCCGTGGGAGCAGAGGTGGGACAGATGCGAGCGTCAGAGGCTGTGGGCGCACTGAGGTTGGGAATGGACCCGAGCAAAGCGATGGCGGAGTCCTTACAAAAGGGGCTGGTCTGTCTGCACGGGAGCTACTTACATTCTAGAGATGGGGATTCCGGAACCCTTGGTGCTAGGTCCACATAGGCCCTGGTTATGGAGTGTGCTCGCTGGCTTCATGG ..........................................................................................((((.(((((...((((.(((.......((((((((((((.....))))))...))))))))).))))..((((((........))).)))..))))).)).))........................................................ ........................................................................................89........................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................TGGGAATGGACCCGAGCAAAGCGATG................................................................................................................................... | 26 | 1 | 10.00 | 10.00 | 5.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .............................................................................................TGGGAATGGACCCGAGCAAAGCGATGG.................................................................................................................................. | 27 | 1 | 8.00 | 8.00 | 2.00 | 2.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGGAATGGACCCGAGCAAAGCGATGGC................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGGCTGTGGGCGCACTGA................................................................................................................................................................ | 18 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TCAGAGGCTGTGGGCGCA.................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGACAGATGCGAGCGT..................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTACATTCTAGAGATGGGGATTCCGGA........................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGCTTTCCAGAGCCTGGGCCACGGCGt......................................................................................................................................................................................................................... | 27 | t | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGCTTTCCAGAGCCTGGGCCACGGCG.......................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGCTTTCCAGAGCCTGGGCCACGGCGGC........................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGACAGATGCGAGCG...................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................................................CAGATGCGAGCGTCAGAGGCTGTGGG........................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTGGGACAGATGCGAGCGTCAGAG................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGCGAGCGTCAGAGGCTGTGGGggca.................................................................................................................................................................... | 26 | ggca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................GAGGCTGTGGGCGCACTGAGGTTG........................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGAGGTTGGGAATGGACCCGAGCt........................................................................................................................................... | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGCGAGCGTCAGAGGCTGTGGGC....................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TAGAGATGGGGATTCCGGAACCCTT..................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGCTACTTACATTCagt........................................................................... | 17 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......TGCTTTCCAGAGCCTGGGCCACGGCGG......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TACAAAAGGGGCTGGTCTGTCTGCA................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CCAGAGCCTGGGCCACGGCGGCGGCCGg.................................................................................................................................................................................................................. | 28 | g | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGGTTATGGAGTGTGCTCGCTGGCTTCgaaa | 31 | gaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................AGGCCCTGGTTATGGAGTGTGCTC............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............CCAGAGCCTGGGCCACGGCGGCGGCCGT.................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TAGAGATGGGGATTCCGGAACCCT...................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGGGGATTCCGGAACCCTTGGTGCTA.............................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................ATGCGAGCGTCAGAGGCTGTGGGCGC..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................ACAAAAGGGGCTGGTCTGTCTGa................................................................................................. | 23 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGACCCGAGCAAAGCGATGGCGGA.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CGGCGGCCGTGGGAGCAGAGGT...................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGAGGTTGGGAATGGACCCGAGCA........................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................ATGGACCCGAGCAAAGCGATGGCGGAG............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGTCTGTCTGCACGGG............................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGACCCGAGCAAAGCGATGGCGGAGT............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGACCCGAGCAAAGCGATGGCGGAGTC........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCGGCTGCTTTCCAGAGCCTGGGCCACGGCGGCGGCCGTGGGAGCAGAGGTGGGACAGATGCGAGCGTCAGAGGCTGTGGGCGCACTGAGGTTGGGAATGGACCCGAGCAAAGCGATGGCGGAGTCCTTACAAAAGGGGCTGGTCTGTCTGCACGGGAGCTACTTACATTCTAGAGATGGGGATTCCGGAACCCTTGGTGCTAGGTCCACATAGGCCCTGGTTATGGAGTGTGCTCGCTGGCTTCATGG ..........................................................................................((((.(((((...((((.(((.......((((((((((((.....))))))...))))))))).))))..((((((........))).)))..))))).)).))........................................................ ........................................................................................89........................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................CCGAGCAAAGCGATGGCGGAGTCCTTA....................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................ATGGACCCGAGCAAAGCGAT.................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................GCAGAGGTGGGACAGATGCGAGCG...................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGACCCGAGCAAAGCGAT.................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................CCACATAGGCCCTGGTTAT........................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CACGGCGGCGGCCGTcac.................................................................................................................................................................................................................. | 18 | cac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................TGGTCTGTCTGCACGGGAGCTACTTACA................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATAGGCCCTGGTTATGGAGTGTGCTC............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |