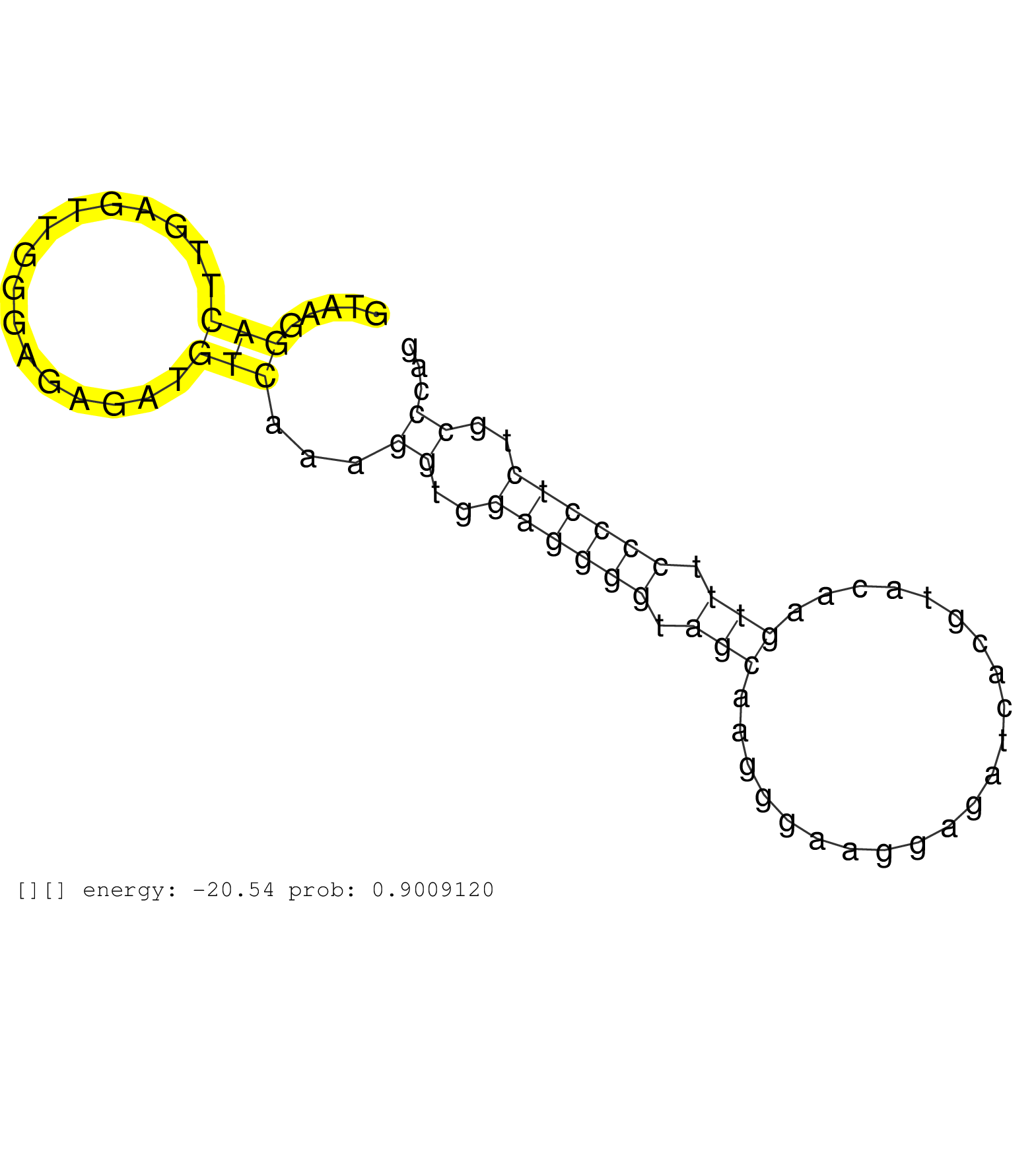

| Gene: Timeless | ID: uc007hlu.1_intron_23_0_chr10_127687519_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(3) PIWI.mut |
(16) TESTES |
| CAGCTCATCTCTCTCTGCTGAAAACCTCGGTGAGAGCCTTCGTCAGGAAGGTAAGGACTTGAGTTGGGAGAGATGTCAAAGGTGGAGGGGTAGCAAGGGAAGGAGATCACGTACAAGTTTCCCCTCTGCCCAGGCCTCTCTGCTCCCCTCCTGTGGCTCCAGAGCTCCCTGATCCGAGCAGCA .......................................................(((................)))...((..((((((.(((......................))).))))))..))..................................................... ..................................................51................................................................................133................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGTC.......................................................................................................... | 27 | 1 | 36.00 | 36.00 | 8.00 | 10.00 | 4.00 | 1.00 | 1.00 | - | 2.00 | 2.00 | 1.00 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | 1.00 |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGT........................................................................................................... | 26 | 1 | 20.00 | 20.00 | 3.00 | 5.00 | 5.00 | 3.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATG............................................................................................................ | 25 | 1 | 10.00 | 10.00 | 4.00 | - | - | - | 1.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ....................................................AAGGACTTGAGTTGGGAGAGATGTCAAA....................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGGACTTGAGTTGGGAGAGATGTC.......................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGTCt......................................................................................................... | 28 | t | 2.00 | 36.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGTCA......................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGGACTTGAGTTGGGAGAGATGT........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACCTCGGTGAGAGCCTTCGTCAGGAAG..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TCCAGAGCTCCCTGATCCGAGCAGCA | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGTt.......................................................................................................... | 27 | t | 1.00 | 20.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGTa.......................................................................................................... | 27 | a | 1.00 | 20.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGGACTTGAGTTGGGAGAGATGTCAAAG...................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGGACTTGAGTTGGGAGAGATGTCA......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGCTGAAAACCTCGGTGAGAGCCTTCGT............................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGACTTGAGTTGGGAGAGATGTtt......................................................................................................... | 28 | tt | 1.00 | 20.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................AGAGCTCCCTGATCCGAG..... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| CAGCTCATCTCTCTCTGCTGAAAACCTCGGTGAGAGCCTTCGTCAGGAAGGTAAGGACTTGAGTTGGGAGAGATGTCAAAGGTGGAGGGGTAGCAAGGGAAGGAGATCACGTACAAGTTTCCCCTCTGCCCAGGCCTCTCTGCTCCCCTCCTGTGGCTCCAGAGCTCCCTGATCCGAGCAGCA .......................................................(((................)))...((..((((((.(((......................))).))))))..))..................................................... ..................................................51................................................................................133................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|