
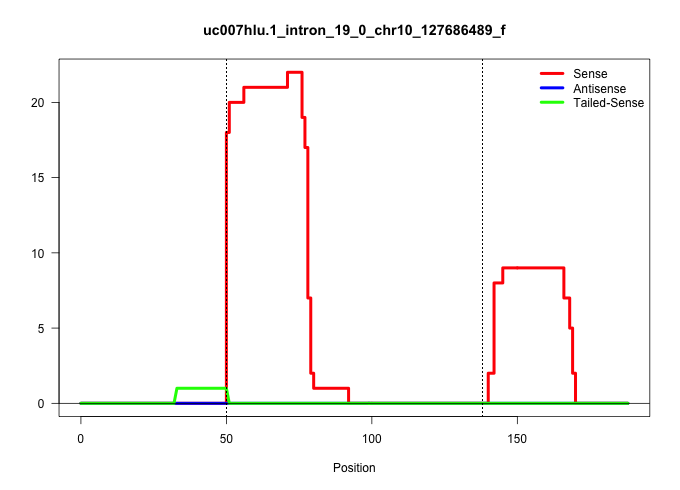
| Gene: Timeless | ID: uc007hlu.1_intron_19_0_chr10_127686489_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| GGAAGAGGCCCAGCTTCAGGAACTATACCTCGCCCACAAGGATGTGGAAGGTGTGTGGTTTAAGATCTCAGGAGACTTTGAGGGGGGCAGTCTGGGCCCCAGACTCCTCCTTACTGCCCCATTCCCTCTCCACCACAGGTCAAGATGTAGTGGAAACCATATTGGCGCACCTGAAAGTCGTTCCTCGA ..................................................(((.((((...........((....))..((((((((((((..(((...........)))..))))))))....)))).))))))).................................................... ..................................................51.....................................................................................138................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGTGTGGTTTAAGATCTCAGGAGACTT.............................................................................................................. | 28 | 1 | 10.00 | 10.00 | 6.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - |
| ...................................ACAAGGATGTGGAAGGTc....................................................................................................................................... | 18 | c | 7.00 | 0.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGATGTAGTGGAAACCATATTGGCGCA................... | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTGTGTGGTTTAAGATCTCAGGAGACTTT............................................................................................................. | 29 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGTGGTTTAAGATCTCAGGAGAC................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................AGATGTAGTGGAAACCATATTGGCGC.................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ...................................................TGTGTGGTTTAAGATCTCAGGAGACTTT............................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGTGGTTTAAGATCTCAGGAGACT............................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................CCACAAGGATGTGGtata......................................................................................................................................... | 18 | tata | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CAAGATGTAGTGGAAACCATATTGGC...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................GAGACTTTGAGGGGGGCAGTC................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGATGTAGTGGAAACCATATTGGC...................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GGTTTAAGATCTCAGGAGACTTTG............................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CAAGATGTAGTGGAAACCATATTGGCGCAC.................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGTAGTGGAAACCATATTGGCGCAC.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GGAAGAGGCCCAGCTTCAGGAACTATACCTCGCCCACAAGGATGTGGAAGGTGTGTGGTTTAAGATCTCAGGAGACTTTGAGGGGGGCAGTCTGGGCCCCAGACTCCTCCTTACTGCCCCATTCCCTCTCCACCACAGGTCAAGATGTAGTGGAAACCATATTGGCGCACCTGAAAGTCGTTCCTCGA ..................................................(((.((((...........((....))..((((((((((((..(((...........)))..))))))))....)))).))))))).................................................... ..................................................51.....................................................................................138................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|