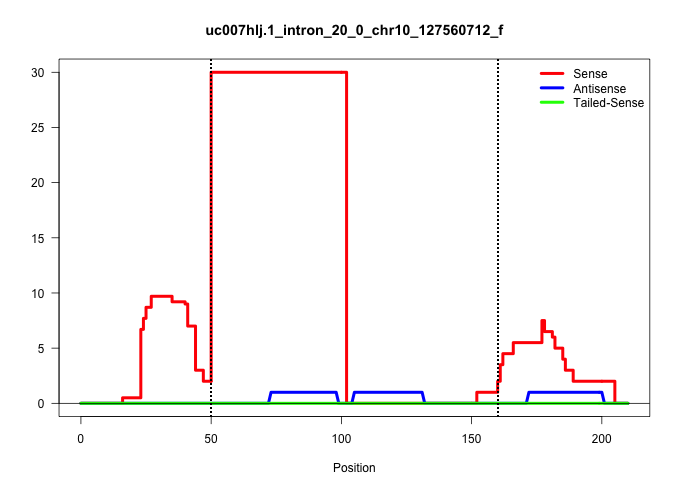
| Gene: Baz2a | ID: uc007hlj.1_intron_20_0_chr10_127560712_f | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(9) TESTES |
| CTGAGGACCAACCTACTCCCTCCCTCCAGCTGCTGGCCTCCTCTAAACCAGTAAGTAGCCAGATCAGTCTGTACTTATCTTGCCCCAGGCTCCCATATCTGGCTGTGAGGCTGTAGCCTGAAGTAGGAAATATGTAAACTCCTTCTTGTCTTCTTTGTAGATGAATACACCCGGTGCTGCCAATCCTTGTTCCCCAGTGCAGCTCTCTTC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTAGCCAGATCAGTCTGTACTTATCTTGCCCCAGGCTCCCATATCTGG............................................................................................................ | 52 | 1 | 30.00 | 30.00 | 30.00 | - | - | - | - | - | - | - | - |
| .......................CTCCAGCTGCTGGCCTCCTCT...................................................................................................................................................................... | 21 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - |
| .......................CTCCAGCTGCTGGCCTCC......................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCCAATCCTTGTTCCCCAGTGCAGCTC..... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - |
| .........................CCAGCTGCTGGCCTCCTCTAAA................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................CTTTGTAGATGAATACACCCGGTGCT................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...........................AGCTGCTGGCCTCCTCTAAACCA................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ACACCCGGTGCTGCCAATCCTTG..................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ........................TCCAGCTGCTGGCCTCCTCT...................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GAATACACCCGGTGCTGCCA............................ | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................ATGAATACACCCGGTGCTGCCAATCC........................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGAATACACCCGGTGCTGCCAATC......................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .......................CTCCAGCTGCTGGCCTCCTCTAAACCA................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGAATACACCCGGTGCTGCC............................. | 20 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - |
| ................TCCCTCCCTCCAGCTGCTG............................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - |
| .......................CTCCAGCTGCTGGCCTC.......................................................................................................................................................................... | 17 | 5 | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - |
| CTGAGGACCAACCTACTCCCTCCCTCCAGCTGCTGGCCTCCTCTAAACCAGTAAGTAGCCAGATCAGTCTGTACTTATCTTGCCCCAGGCTCCCATATCTGGCTGTGAGGCTGTAGCCTGAAGTAGGAAATATGTAAACTCCTTCTTGTCTTCTTTGTAGATGAATACACCCGGTGCTGCCAATCCTTGTTCCCCAGTGCAGCTCTCTTC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....CAACCTACTCCCTCCtacg........................................................................................................................................................................................... | 19 | tacg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................CCCAGTGCAGCTCTCc... | 16 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................TGAGGCTGTAGCCTGAAGTAGGAAATA.............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .....CAACCTACTCCCTCCtat........................................................................................................................................................................................... | 18 | tat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................CTTATCTTGCCCCAGGCTCCCATATC............................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................GGTGCTGCCAATCCTTGTTCCCCAGTGCA......... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |