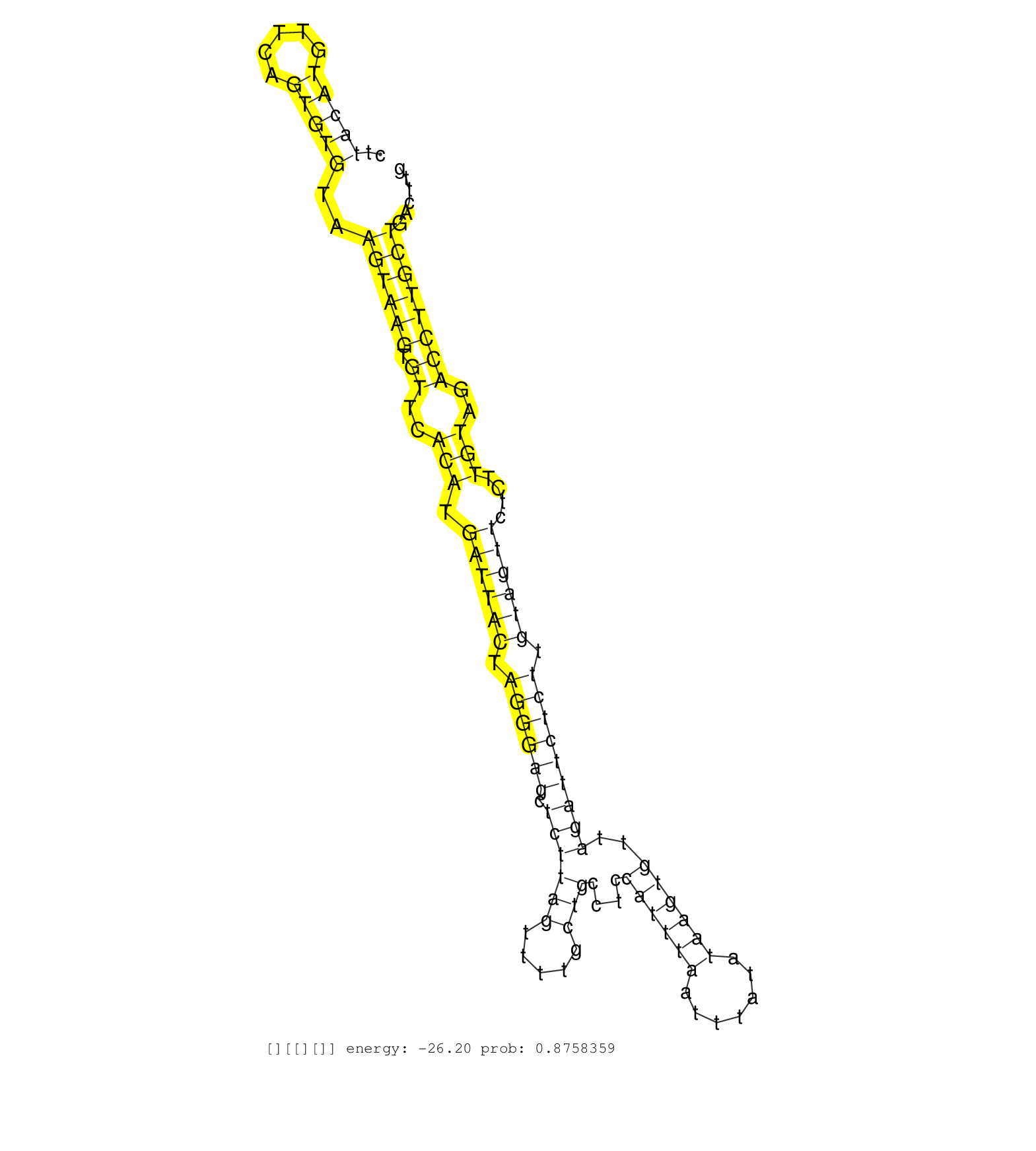
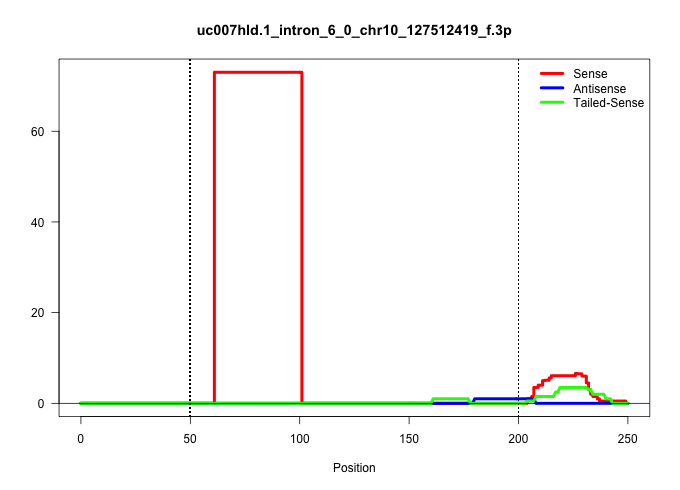
| Gene: Ptges3 | ID: uc007hld.1_intron_6_0_chr10_127512419_f.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(20) TESTES |
| ACTAAGAACCAAAAAGTCTTGTCACATTCTTAGGAGGGTTTATTATTTAAGGTGTGCTTACATGTTCAGTGTGTAAGTAAGTGTTCACATGATTACTAGGGAGCTCTTAGTTTTGCTGCCTCCATTTAATTTATATAAGTGTTAGATTCTCTTGTAGTTCTCTTGTAGACCTTGCTGACTTGTCTGTAAAATTCTTTCAGAGATGCCAGATCTGGAGTAAGGGCTGTTGTCATCGCCTGGATTTTGAGAA ..........................................................(((((.....)))))..((((((.((..(((.((((((.((((((.((((((.....)))....((((((.......))))))..))))))))).))))))....)))..)))))))).......................................................................... ........................................................57...........................................................................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................ATGTTCAGTGTGTAAGTAAGTGTTCACATGATTACTAGGGAGCTCTTAGTTT......................................................................................................................................... | 52 | 1 | 74.00 | 74.00 | 74.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GGAGTAAGGGCTGTTGTC................... | 18 | 2 | 3.50 | 3.50 | - | 1.00 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................ATCTGGAGTAAGGGCTGTTGTCATCGC.............. | 27 | 2 | 1.50 | 1.50 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...............................................................................................................................................................................................................AGATCTGGAGTAAGGGCTGTTGTCATC................ | 27 | 2 | 1.50 | 1.50 | - | - | - | - | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TAAGGGCTGTTGTCATCGCCTGt.......... | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGATCTGGAGTAAGGGCTGTTGTC................... | 24 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................CTTGTAGACCTTGtttc........................................................................ | 17 | tttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GAGTAAGGGCTGTTGTCATCGC.............. | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGGGCTGTTGTCATCGCCTGGATg....... | 24 | g | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GATCTGGAGTAAGGGCTGTTGTCATt................ | 26 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GGAGTAAGGGCTGTTGTCAT................. | 20 | 2 | 1.00 | 1.00 | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AGTAAGGGCTGTTGTC................... | 16 | 3 | 0.67 | 0.67 | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GTTGTCATCGCCTGGATTTTGA... | 22 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................CTGGAGTAAGGGCTGTTGTCA.................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GAGTAAGGGCTGTTGTCA.................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GCCAGATCTGGAGTAAGGGCTGTTGTC................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGGAGTAAGGGCTGTTG..................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................CTGGAGTAAGGGCTGTTGTCAT................. | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................ATCTGGAGTAAGGGCTGTTG..................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TTGTCATCGCCTGGATTTTGAGA. | 23 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGATCTGGAGTAAGGGCTGTTGTCAT................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................CTGGAGTAAGGGCTGTTGTCATC................ | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ..............................................................................................................................................................................................................CAGATCTGGAGTAAGGGCTGTTGTCA.................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GCCAGATCTGGAGTAAGGGCTGTTGTCt.................. | 28 | t | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .........................................................................................................................................................................................................................TAAGGGCTGTTGTCATCG............... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| .......................................................................................................................................................................................................................AGTAAGGGCTGTTGTCATCGCC............. | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AGTAAGGGCTGTTGTCATCGCCTGGATT....... | 28 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GCCAGATCTGGAGTAAGGGCTGTTG..................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ....................................................................................................................................................................................................................TGGAGTAAGGGCTGT....................... | 15 | 12 | 0.08 | 0.08 | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACTAAGAACCAAAAAGTCTTGTCACATTCTTAGGAGGGTTTATTATTTAAGGTGTGCTTACATGTTCAGTGTGTAAGTAAGTGTTCACATGATTACTAGGGAGCTCTTAGTTTTGCTGCCTCCATTTAATTTATATAAGTGTTAGATTCTCTTGTAGTTCTCTTGTAGACCTTGCTGACTTGTCTGTAAAATTCTTTCAGAGATGCCAGATCTGGAGTAAGGGCTGTTGTCATCGCCTGGATTTTGAGAA ..........................................................(((((.....)))))..((((((.((..(((.((((((.((((((.((((((.....)))....((((((.......))))))..))))))))).))))))....)))..)))))))).......................................................................... ........................................................57...........................................................................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TGCTGCCTCCATTTAAgt......................................................................................................................... | 18 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................TGTCTGTAAAATTCTTTCAGAGATGCCA.......................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |