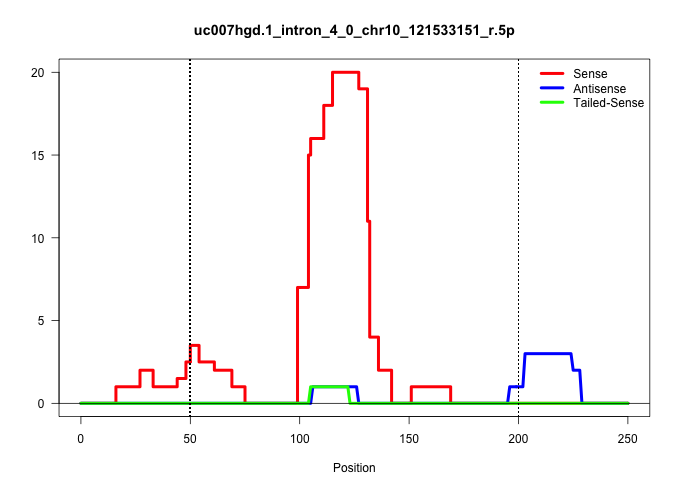
| Gene: Tmem5 | ID: uc007hgd.1_intron_4_0_chr10_121533151_r.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(1) PIWI.mut |
(13) TESTES |
| GAACTCCAGGAAGGCGGCGGCGGCGCAGGCGAAGGAGAGGCGCGGCCGAGGTAGGACGCGGGCCGCACGGGGCAGGGGCCTAGCGGCGCTGTCGTGTGCGTGGACGCCGGGCGGCAGGCACTGCGCAGACGAGCCACGCGGGCCTCGGCTCGGGCCTCGGCTCTCTCGGGGTCCTCAGCCCCGGCAGCCAGCTCTGGGCTCGGCTCGGAGGCTCTGCTCGCCCTGGGCGTGCACGCAGGCCTTTAAAACG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................CGCCGGGCGGCAGGCACTGCGCAGACG....................................................................................................................... | 27 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GTGGACGCCGGGCGGCAGGCACTGCGCAGACGA...................................................................................................................... | 33 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CGGCAGGCACTGCGCAGACGAGCCACGCGGG............................................................................................................ | 31 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AGGCACTGCGCAGACGAGCCA.................................................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................GCCGGGCGGCAGGCACTt............................................................................................................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................GGCGAAGGAGAGGCGCGGCCGAGGTAG.................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAGGACGCGGGCCGCACG..................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................AGGTAGGACGCGGGCCGCACGGGGCAG............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................................GGGCCTCGGCTCTCTCGG................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................GCCGGGCGGCAGGCACTGCGCA........................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................GCGGCGGCGCAGGCGAA......................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................GCCGAGGTAGGACGCGG............................................................................................................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| GAACTCCAGGAAGGCGGCGGCGGCGCAGGCGAAGGAGAGGCGCGGCCGAGGTAGGACGCGGGCCGCACGGGGCAGGGGCCTAGCGGCGCTGTCGTGTGCGTGGACGCCGGGCGGCAGGCACTGCGCAGACGAGCCACGCGGGCCTCGGCTCGGGCCTCGGCTCTCTCGGGGTCCTCAGCCCCGGCAGCCAGCTCTGGGCTCGGCTCGGAGGCTCTGCTCGCCCTGGGCGTGCACGCAGGCCTTTAAAACG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................CTCGGAGGCTCTGCTCGCCCTGGGCG..................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CCGGGCGGCAGGCACTGCGCA........................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................................GGCTCGGCTCGGAGGCTCTGCTCGCCCTG......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |