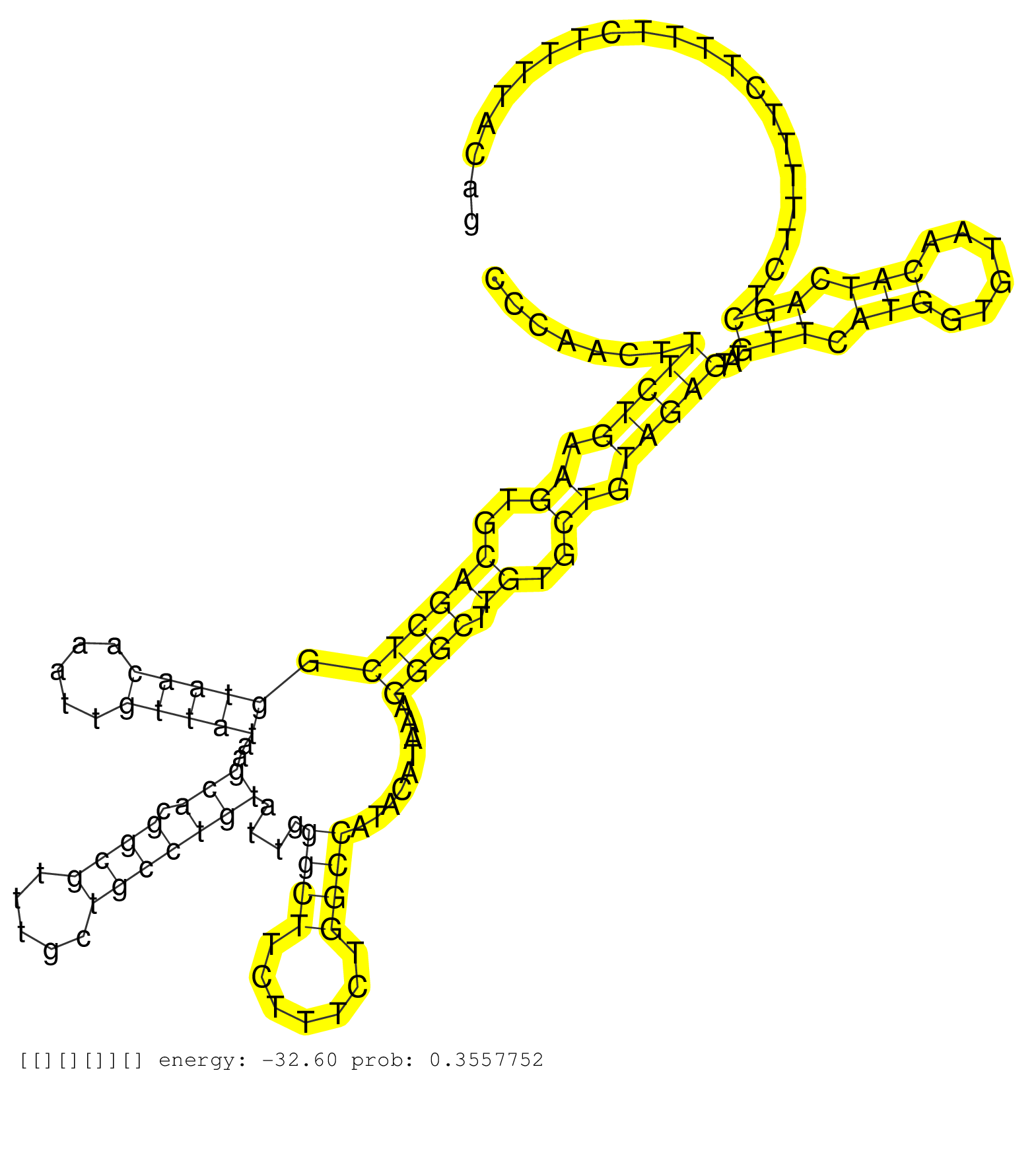

| Gene: Mdm2 | ID: uc007hdl.1_intron_7_0_chr10_117142286_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| GCATTTTCTGTCCTAACGCTTAGTGGTGTGCACTTGTACTTAGTGTAAAACCCAACTTTCTGAAGTGCAGCTCGGTAACAAATTGTTATAAGCACGGCGTTTGCTGCCTGTATTGGGCTTCTTTCTGGCCATACATAAAGGGCTTGTGCTGTAGAGTATGTTCATGGTGTAACATCAGCTCTTTTTCTTTTCTTTTACAGGTTAGACCAAAACCATTGCTTTTGAAGTTGTTAAAGTCCGTTGGAGCGCA .........................................................(((((.((..((((((.(((((.....)))))..(((.((((.....)))))))....((((.......)))).........)))).))..)).)))))...(((.(((......))).)))....................................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................CTTCTTTCTGGCCATACATAAAGGGCTTGTGCTGTAGAGTATGTTCATGGTG................................................................................. | 52 | 1 | 166.00 | 166.00 | 166.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGTGCTGTAGAGTATGTTCA...................................................................................... | 20 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAAGTTGTTAAAGTCCGTTGGAGCGC. | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAAGTTGTTAAAGTCCGTTGGAGCGCA | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAAGTTGTTAAAGTCCGTTGG...... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GAAGTTGTTAAAGTCCGTTGGAGCGCA | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GAAGTTGTTAAAGTCCGTTGGAGCGC. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TTGTTAAAGTCCGTTGGAGCGC. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................................TGCTTTTGAAGTTGTTAAAGTCCGTTGGA..... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................AAAACCCAACTTTCTGAAGTGCAGCTCG................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................GAAGTGCAGCTCGGTAACAAATTG..................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AAAACCATTGCTTTTGAAGTTGTT.................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................AAAGGGCTTGTGCTGTAGAGTATGTTCA...................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| GCATTTTCTGTCCTAACGCTTAGTGGTGTGCACTTGTACTTAGTGTAAAACCCAACTTTCTGAAGTGCAGCTCGGTAACAAATTGTTATAAGCACGGCGTTTGCTGCCTGTATTGGGCTTCTTTCTGGCCATACATAAAGGGCTTGTGCTGTAGAGTATGTTCATGGTGTAACATCAGCTCTTTTTCTTTTCTTTTACAGGTTAGACCAAAACCATTGCTTTTGAAGTTGTTAAAGTCCGTTGGAGCGCA .........................................................(((((.((..((((((.(((((.....)))))..(((.((((.....)))))))....((((.......)))).........)))).))..)).)))))...(((.(((......))).)))....................................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................CCATTGCTTTTGAAGcgt....................... | 18 | cgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |