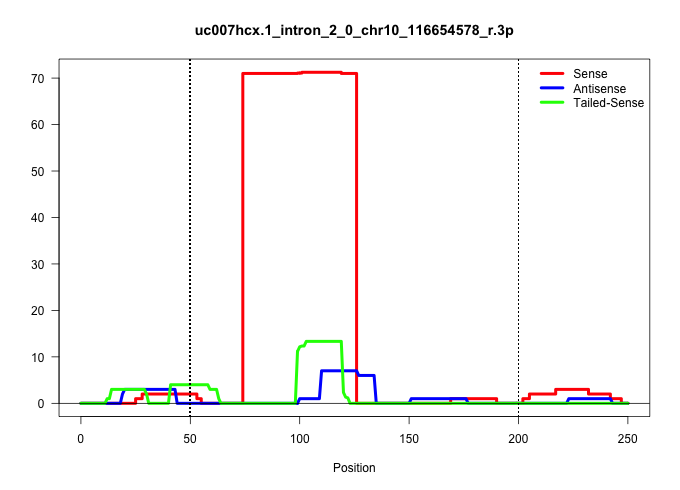
| Gene: Yeats4 | ID: uc007hcx.1_intron_2_0_chr10_116654578_r.3p | SPECIES: mm9 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(23) TESTES |
| GTCAGGCGAGACTGGTGGCGCACGCTTGTAACCTTAGCCCTTGAGAAGTGGAGGCAGGAAGGTCAGCCAGAAGTTCAAGGCCTGCCTTCTGTTACTTAGTGAGTTCGAGGCCAGCCTCCAGGTCTCACAAGTCCAGGAAGGACTGACTCACTGACAGAAGTGTTCTGACCGTGGATAGAGTGACCCTGTGTGTGTTCTAGGTGACTCTGTACCACTTACTGAAGCTCTTCCAGTCTGACACCAATGCCAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TCAAGGCCTGCCTTCTGTTACTTAGTGAGTTCGAGGCCAGCCTCCAGGTCTC............................................................................................................................ | 52 | 1 | 71.00 | 71.00 | 71.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGAGTTCGAGGCCAGCCgctc.................................................................................................................................. | 21 | gctc | 11.00 | 0.00 | - | 5.00 | - | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............GTGGCGCACGCTTGTAg........................................................................................................................................................................................................................... | 17 | g | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TGAGAAGTGGAGGCAGGAAGGa........................................................................................................................................................................................... | 22 | a | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................ACTGAAGCTCTTCCAGTCTGACACC........ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................TTCGAGGCCAGCCTCCttac............................................................................................................................... | 20 | ttac | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCTGTACCACTTACTGAAGCTCTTCCA.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................TGAGAAGTGGAGGCgggg............................................................................................................................................................................................... | 18 | gggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................................................GACTCTGTACCACTTACTGAAGCTCT...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................TAACCTTAGCCCTTGAGAAGTGGAGGC................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTGTAACCTTAGCCCTTGAGAAGTGGAG..................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGGTGGCGCACGCTTctt............................................................................................................................................................................................................................ | 18 | ctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................................................................TCCAGTCTGACACCAATGC... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................GAGTTCGAGGCCAGCCTCCgc................................................................................................................................. | 21 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................TGAGAAGTGGAGGCAGGAAGaaa.......................................................................................................................................................................................... | 23 | aaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CGTGGATAGAGTGACCCTGTG............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AGTTCGAGGCCAGCCTCC................................................................................................................................... | 18 | 12 | 0.17 | 0.17 | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGAGTTCGAGGCCAGCCTCCttaa............................................................................................................................... | 24 | ttaa | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 |
| ...................................................................................................TGAGTTCGAGGCCAGCCTCCtaa................................................................................................................................ | 23 | taa | 0.08 | 0.08 | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AGTTCGAGGCCAGCCTCCtcc................................................................................................................................ | 21 | tcc | 0.08 | 0.17 | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AGTTCGAGGCCAGCCTCCtctt............................................................................................................................... | 22 | tctt | 0.08 | 0.17 | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGAGTTCGAGGCCAGCCTCC................................................................................................................................... | 20 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTCAGGCGAGACTGGTGGCGCACGCTTGTAACCTTAGCCCTTGAGAAGTGGAGGCAGGAAGGTCAGCCAGAAGTTCAAGGCCTGCCTTCTGTTACTTAGTGAGTTCGAGGCCAGCCTCCAGGTCTCACAAGTCCAGGAAGGACTGACTCACTGACAGAAGTGTTCTGACCGTGGATAGAGTGACCCTGTGTGTGTTCTAGGTGACTCTGTACCACTTACTGAAGCTCTTCCAGTCTGACACCAATGCCAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................CCAGCCTCCAGGTCTCACAAGTCCA................................................................................................................... | 25 | 1 | 6.00 | 6.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GCACGCTTGTAACCTTAGCCCTTGA.............................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TGACAGAAGTGTTCTGACCGTGGATA......................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................CACGCTTGTAACCTTAGCCCTTGA.............................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................GAGTTCGAGGCCAGCCTCCggt................................................................................................................................... | 22 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................GAGTTCGAGGCCAGCCTCCAGGTCTCA........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................AAGGTCAGCCAGAAGTaga................................................................................................................................................................................ | 19 | aga | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGTTCGAGGCCAGCCTCCgt................................................................................................................................... | 22 | gt | 1.00 | 0.08 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GCTCTTCCAGTCTGACACCA....... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |