
| Gene: Tbc1d15 | ID: uc007hax.1_intron_0_0_chr10_114636821_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(13) TESTES |
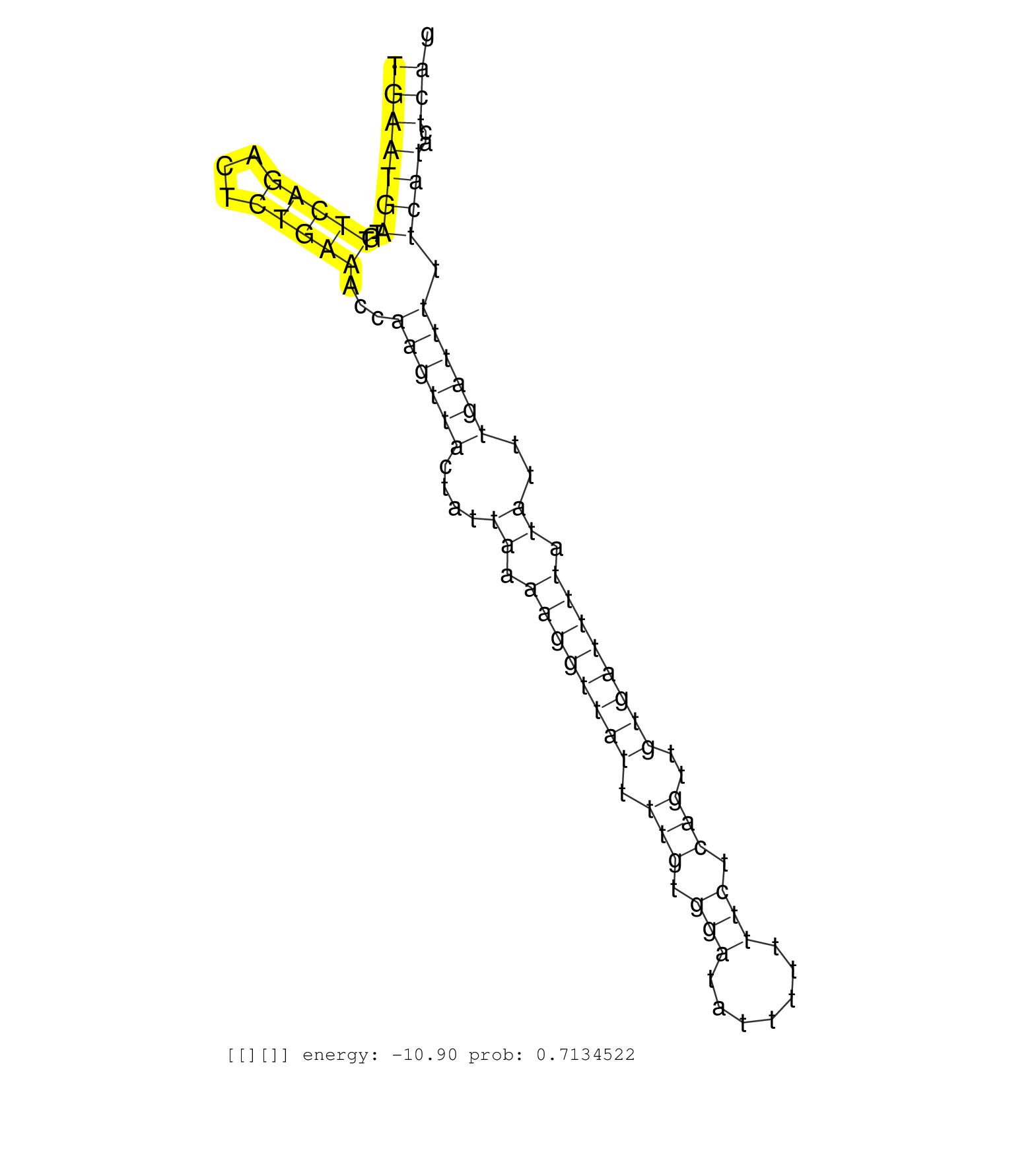
| TACCTCAGTGGGGATGCCCCAGTGTGGTCTGCTAAGGCACCTCCTCCCGCTGCATCTTGCTGCCTCGTTACAGAACATATCAGCACTTGCCCAGTTCAAATGAATGATGTTCAGACTCTGAAACCAAGTTACTATTAAAAGGTTATTTTGTGGATATTTTTTTCTCAGTTGTGATTTTATATTTGATTTTTCATACTCAGGAATTGCCACAAGCAGTTTGTGAGATCCTGGGACTTCAGGACAGTGAAAT ....................................................................................................(((((((..(((((...)))))...((((((....((.((((((((.(((.(((.......))).)))..)))))))).))..)))))).))))..)))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................ATGAATGATGTTCAGACTCTGAAA............................................................................................................................... | 24 | 1 | 17.00 | 17.00 | 10.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AATGAATGATGTTCAGACTCTGAAA............................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................................CTGGGACTTCAGGACAG...... | 17 | 2 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................ATGAATGATGTTCAGACTCTGAA................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ACAAGCAGTTTGTGAGATCCTGGGA................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................CACAAGCAGTTTGTGAGATCCTGGG.................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGAGATCCTGGGACTTCAGGACAGTGA... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................TGAAACCAAGTTACTtaaa................................................................................................................. | 19 | taaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................ACAAGCAGTTTGTGAGATCCTGGGAC................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................GGGACTTCAGGACAGTGA... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................ATGAATGATGTTCAGACTCTGAAAa.............................................................................................................................. | 25 | a | 1.00 | 17.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................CACAAGCAGTTTGTGAGATCCTGGGA................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| TACCTCAGTGGGGATGCCCCAGTGTGGTCTGCTAAGGCACCTCCTCCCGCTGCATCTTGCTGCCTCGTTACAGAACATATCAGCACTTGCCCAGTTCAAATGAATGATGTTCAGACTCTGAAACCAAGTTACTATTAAAAGGTTATTTTGTGGATATTTTTTTCTCAGTTGTGATTTTATATTTGATTTTTCATACTCAGGAATTGCCACAAGCAGTTTGTGAGATCCTGGGACTTCAGGACAGTGAAAT ....................................................................................................(((((((..(((((...)))))...((((((....((.((((((((.(((.(((.......))).)))..)))))))).))..)))))).))))..)))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................AATTGCCACAAGCAGtttg.................................. | 19 | tttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |