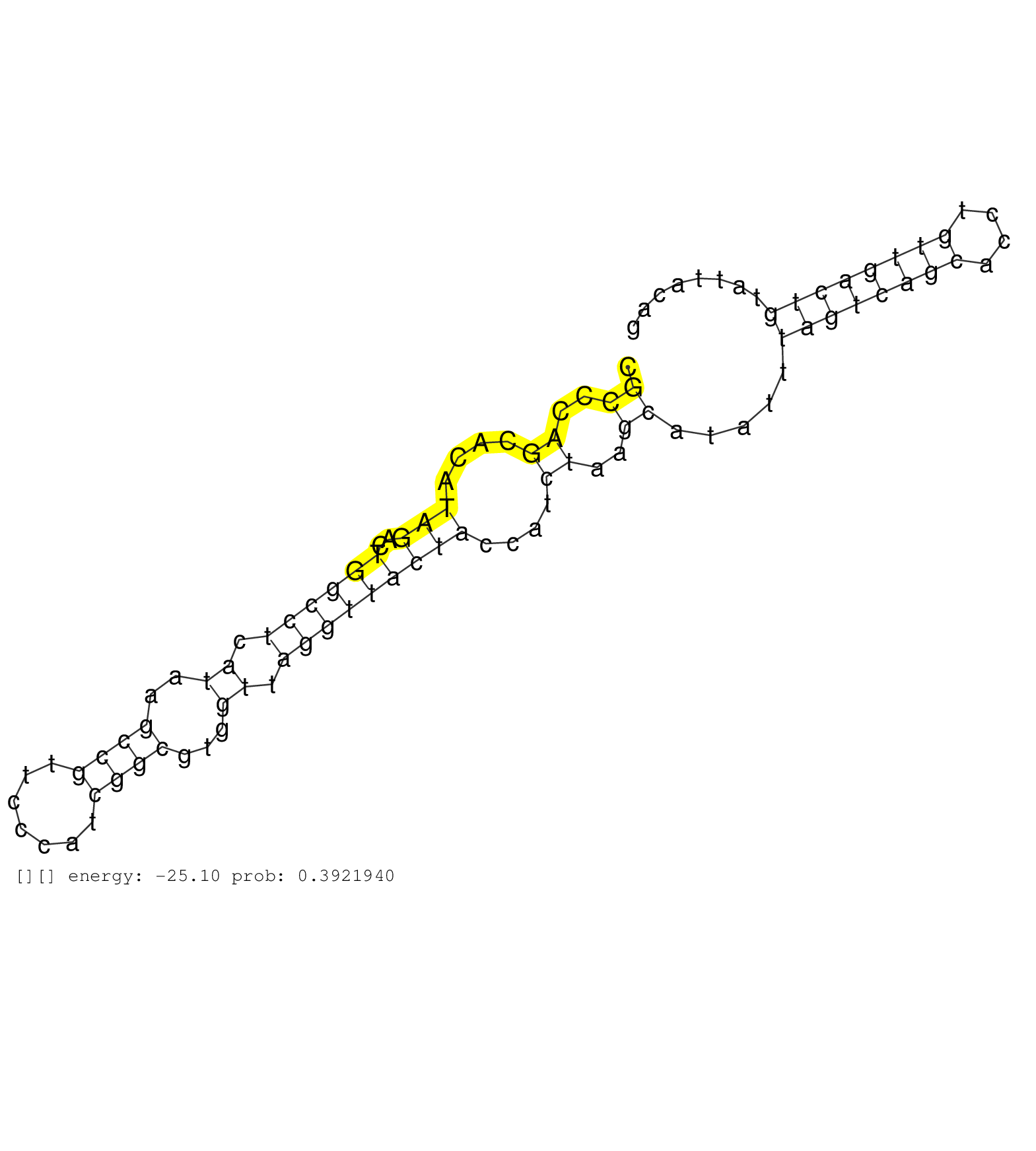

| Gene: Pctk2 | ID: uc007gul.1_intron_8_0_chr10_92689521_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(12) TESTES |
| AGCAATACATGGATGACTGTGGGAACATCATGAGCATGCACAATGTCAAGGTATGTTGTCCATGCACTTCTGAGCCTGTGTCAGCACAGCTGGACTGGATCGCCCAGCACATAGACTGGCCTCATAAGCCGTTCCCATCGGCGTGGTTAGGTTACTACCATCTAAGCATATTTAGTCAGCACCTGTTGACTGTATTACAGCTGTTAAAAAACCAGACCCTTTGGAGTGGTGCTTTGCTTTAAAGGTCTTT .....................................................................................................((..((....(((..((((((.((..((((.......))))...)).)))))))))....))..)).....((((((((....)))))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATGTTGTCCATGCACTTCTGAGCCTGTGTCAGCACAGCTGGACTGGATCG.................................................................................................................................................... | 52 | 1 | 28.00 | 28.00 | 28.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......CATGGATGACTGTGGGAACAT.............................................................................................................................................................................................................................. | 21 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - |
| ..............GACTGTGGGAACATCATGAGCATGCACAATGT............................................................................................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............TGACTGTGGGAACATCATGAGC....................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CTTTGGAGTGGTGCgcgt.............. | 18 | gcgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................TGGGAACATCATGAGCATGCACAATGTCAt......................................................................................................................................................................................................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................TGGGAACATCATGAGCATGCACAATGTC........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........GATGACTGTGGGAACATCATGAGCATGCA.................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....ATACATGGATGACTGTGGGg.................................................................................................................................................................................................................................. | 20 | g | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........ATGGATGACTGTGGGAACATCATGAGCATGC................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................CAGCACAGCTGGACTGGATCGC................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....ATACATGGATGACTGTGGGAACA............................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................................................................GTGGTGCTTTGCTTTcgca...... | 19 | cgca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| AGCAATACATGGATGACTGTGGGAACATCATGAGCATGCACAATGTCAAGGTATGTTGTCCATGCACTTCTGAGCCTGTGTCAGCACAGCTGGACTGGATCGCCCAGCACATAGACTGGCCTCATAAGCCGTTCCCATCGGCGTGGTTAGGTTACTACCATCTAAGCATATTTAGTCAGCACCTGTTGACTGTATTACAGCTGTTAAAAAACCAGACCCTTTGGAGTGGTGCTTTGCTTTAAAGGTCTTT .....................................................................................................((..((....(((..((((((.((..((((.......))))...)).)))))))))....))..)).....((((((((....)))))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|