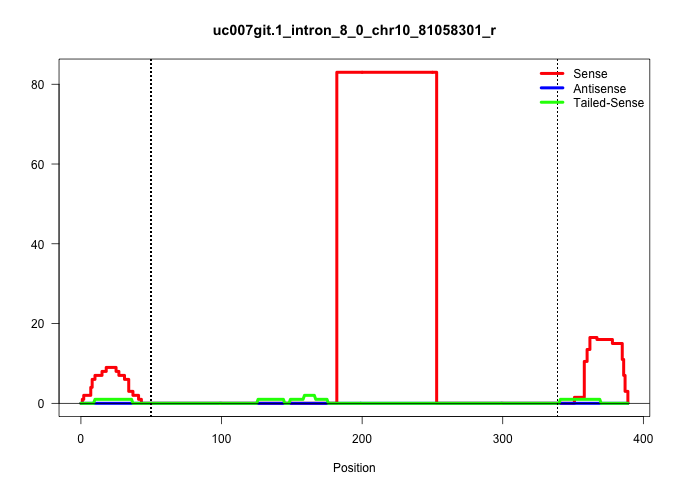
| Gene: Tle6 | ID: uc007git.1_intron_8_0_chr10_81058301_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(23) TESTES |
| ACAGCCGCCAAAAGAACCAACAGGAACACGGGGGTGAGGATTCCTCACAGGTGAGGGGGAACTCCCAGGCACAAGCCTTTCTCTCCCTCTTCTTCCCTCTCTCCTCCCCCCACCTTGCCTGTTGTCATGGCAACAGAGAAGCTGCTGGAATTGGTGAGTACCCTTGACTGGAACCCCAAAGGATTTTACCCCATAGTGTCCCCTCAGTACCCTCCTTCCTCATCCCTGCACTCTGACCCCTCAGGAACATCCCCACTGCAGCCAGGAACCCCCAGGCATCTGCTGGGTGTCCCCCCCTGGGGTGACGTGTCCCCCATCTGACCCAATTCTGTCTCCCAGGAAAGCAAAGATTCAGGCCTGTGTGACTTTAAACCAGAACCTCAGCCCAG ...................................................................................................................................................................................................................................................((.((((((.....((((((.((.....)).)))...))).)))))).))....(((((.((....)))))))...(((........)))........................................................ ...............................................................................................................................................................................................................................................240................................................................................................339................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................ATTTTACCCCATAGTGTCCCCTCAGTACCCTCCTTCCTCATCCCTGCACTCT........................................................................................................................................................... | 52 | 1 | 85.00 | 85.00 | 85.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................TGTGTGACTTTAAACCAGAACCTCAGCC... | 28 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................TGTGTGACTTTAAACCAGAACCTCAGC.... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................TGTGACTTTAAACCAGAACCTCAGCCC.. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........CAAAAGAACCAACAGGAACACGGGGG................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................TGTGTGACTTTAAACCAGAACCTCAGCCC.. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................TGACTTTAAACCAGAACCTCAGCCCAG | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................TGTGACTTTAAACCAGAACCTCAGCCCAG | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........AAAGAACCAACAGGAACACGGGGGTGA................................................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................ATGGCAACAGAGAAGtcca.................................................................................................................................................................................................................................................... | 19 | tcca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................TGACTTTAAACCAGAACCTCAGC.... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................AAGCAAAGATTCAGGCCTGTGTGACTTTt................... | 29 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................ATTGGTGAGTACCCcgcc.............................................................................................................................................................................................................................. | 18 | cgcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................GGAAAGCAAAGATTCAGGCCTGTGT.......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AACAGGAACACGGGGGTGAGGATTC.......................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............ACCAACAGGAACACGGGGGTGAGGAT............................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................ACCCTTGACTGGAAgct..................................................................................................................................................................................................................... | 17 | gct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CCAAAAGAACCAACAGGAACACGG...................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................TGTGACTTTAAACCAGAACCTCAGC.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .CAGCCGCCAAAAGAACCAACAGGAAC.......................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................TCAGGCCTGTGTGACTTTAAACCAGAA........... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CCAAAAGAACCAACAGGAACACGGGGG................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..AGCCGCCAAAAGAACCAACAGGA............................................................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................................................................................AGGAAAGCAAAGATTCAGGCCTGTGTGA........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AAAGAACCAACAGGAACACGGGGGTta................................................................................................................................................................................................................................................................................................................................................................ | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................TCAGGCCTGTGTGACT...................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ACAGCCGCCAAAAGAACCAACAGGAACACGGGGGTGAGGATTCCTCACAGGTGAGGGGGAACTCCCAGGCACAAGCCTTTCTCTCCCTCTTCTTCCCTCTCTCCTCCCCCCACCTTGCCTGTTGTCATGGCAACAGAGAAGCTGCTGGAATTGGTGAGTACCCTTGACTGGAACCCCAAAGGATTTTACCCCATAGTGTCCCCTCAGTACCCTCCTTCCTCATCCCTGCACTCTGACCCCTCAGGAACATCCCCACTGCAGCCAGGAACCCCCAGGCATCTGCTGGGTGTCCCCCCCTGGGGTGACGTGTCCCCCATCTGACCCAATTCTGTCTCCCAGGAAAGCAAAGATTCAGGCCTGTGTGACTTTAAACCAGAACCTCAGCCCAG ...................................................................................................................................................................................................................................................((.((((((.....((((((.((.....)).)))...))).)))))).))....(((((.((....)))))))...(((........)))........................................................ ...............................................................................................................................................................................................................................................240................................................................................................339................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................................................................................GCAAAGATTCAGGCCTGTGTGACTTTA................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CATCCCTGCACTCTcgaa........................................................................................................................................................... | 18 | cgaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CATCCCTGCACTCTcaaa........................................................................................................................................................... | 18 | caaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |