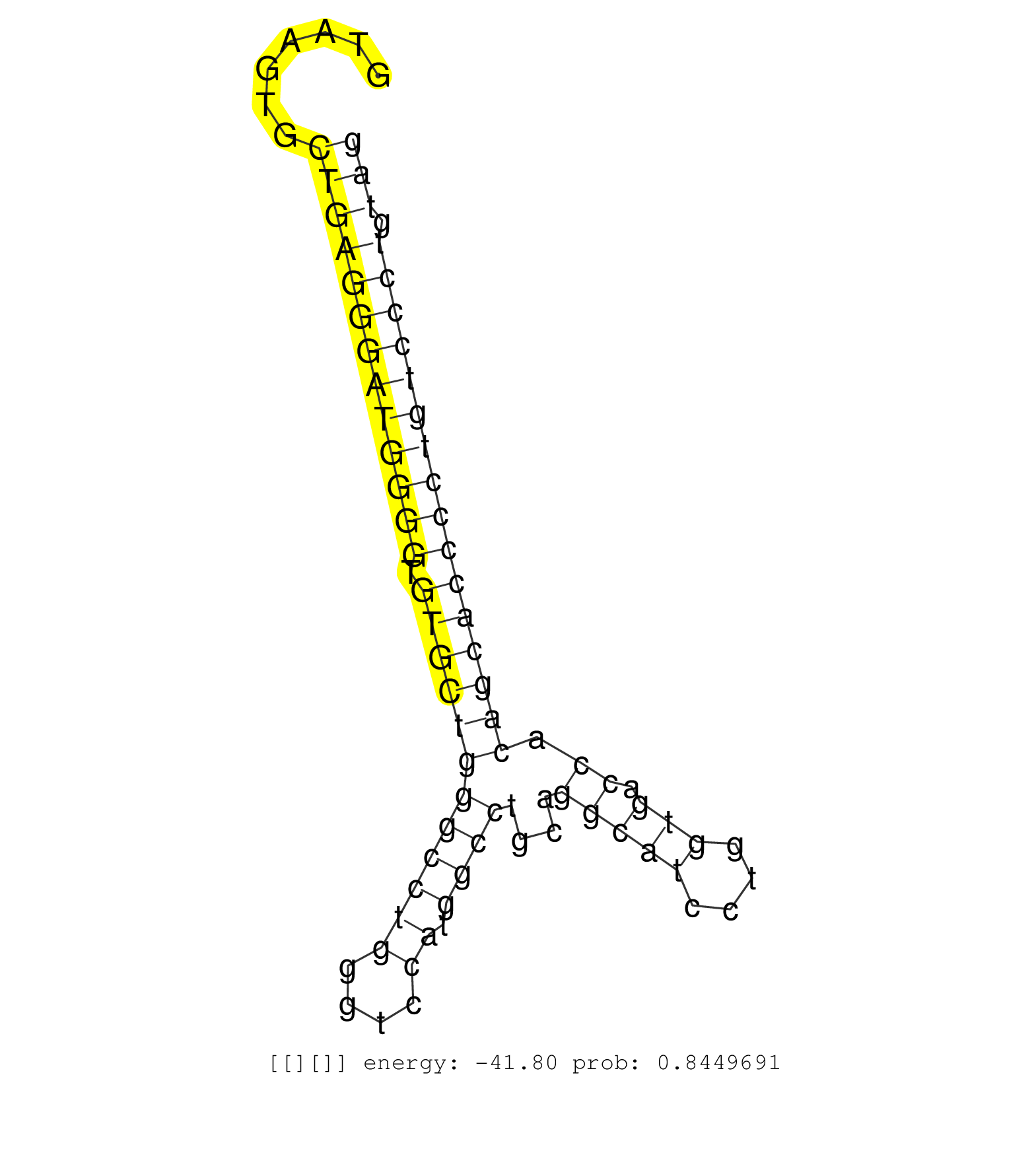

| Gene: Ap3d1 | ID: uc007gek.1_intron_23_0_chr10_80186352_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(25) TESTES |
| TCATGACATCCTCCACCAACAACTGGGTCCTCATCAAGATCATCAAGCTGGTAAGTGCTGAGGGATGGGGTGTGCTGGGCCTGGGTCCATGGCCTGCAGGCATCCTGGTGACCACAGCACCCCTGTCCCTGTAGTTCGGTGCCCTGACTCCTCTGGAGCCCAGGCTGGGCAAGAAGCTGATCGA .........................................................(((((((((((((.((((((((((((....)).))))....(((((....))).)).)))))))))))))))).))).................................................. ..................................................51.................................................................................134................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGCTGAGGGATGGGGTGTGC............................................................................................................. | 25 | 1 | 46.00 | 46.00 | 16.00 | 1.00 | 8.00 | - | 5.00 | 4.00 | 1.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTG.............................................................................................................. | 24 | 1 | 11.00 | 11.00 | 2.00 | 2.00 | - | - | - | - | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTGa............................................................................................................. | 25 | a | 6.00 | 11.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTGCT............................................................................................................ | 26 | 1 | 5.00 | 5.00 | - | 1.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGG.................................................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTGt............................................................................................................. | 25 | t | 3.00 | 11.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTGCTG........................................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGT............................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTG................................................................................................................ | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGGATGGGGTGTGCTGGGC........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CTGGGTCCTCATCAAGATCATCAAGCTG...................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGggtg.............................................................................................................. | 24 | ggtg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGCTGAGGGATGGGGTG................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................ATCAAGATCATCAAGCTGGTA................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGcgg............................................................................................................... | 23 | cgg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGCTGAGGGATGGGGTGTGCT............................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGgg.............................................................................................................. | 24 | gg | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTGCTGAGGGATGGGGTGTGCTGt.......................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGcg................................................................................................................ | 22 | cg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGCTGAGGGATGGGGTGTGCTGGGC........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTGg............................................................................................................. | 25 | g | 1.00 | 11.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGCTGAGGGATGGGGcggg.............................................................................................................. | 21 | cggg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGTtc............................................................................................................. | 25 | tc | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCCAGGCTGGGCAAGAAGCTGATC.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGggc............................................................................................................. | 25 | ggc | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CTGAGGGATGGGGTGTGCTGGGCC....................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGTGCTGAGGGATGGGGTGaga............................................................................................................. | 25 | aga | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGCTGAGGGATGGGGTGTGCTGGGCC....................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CACCCCTGTCCCTGTAGTTCGGTGCC......................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TCAAGATCATCAAGCTGttc................................................................................................................................... | 20 | ttc | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .................................TCAAGATCATCAAGCTG...................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCATGACATCCTCCACCAACAACTGGGTCCTCATCAAGATCATCAAGCTGGTAAGTGCTGAGGGATGGGGTGTGCTGGGCCTGGGTCCATGGCCTGCAGGCATCCTGGTGACCACAGCACCCCTGTCCCTGTAGTTCGGTGCCCTGACTCCTCTGGAGCCCAGGCTGGGCAAGAAGCTGATCGA .........................................................(((((((((((((.((((((((((((....)).))))....(((((....))).)).)))))))))))))))).))).................................................. ..................................................51.................................................................................134................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................GTGACCACAGCACCCtgc.............................................................. | 18 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |