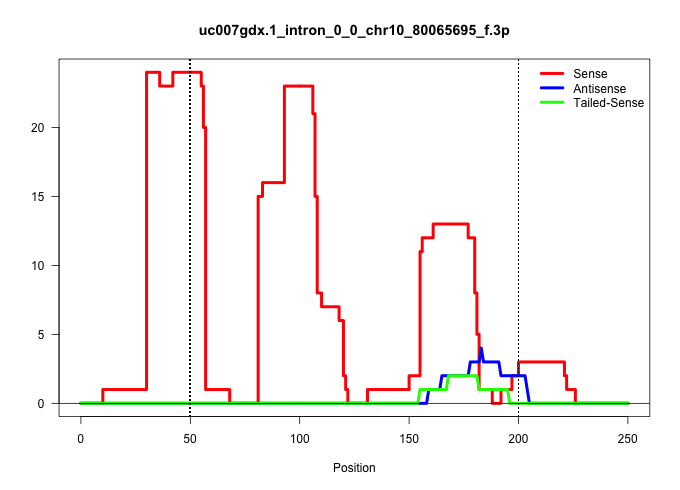
| Gene: Adat3 | ID: uc007gdx.1_intron_0_0_chr10_80065695_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(2) PIWI.mut |
(24) TESTES |
| TGTTGGGACCACGTGGCCTCGTCAGAGCCCTGAAGAGCCCGGCACGATTATTCCTTATGCGGCCGGGGAATAAGCCAACTCTACCTGGCGCTCTGGACTCCTCATTCCTGGGCCTGGCGGGACCCACCACATCCCAGGACAGACCCCCGCCCCACTGAAGCTGACGGATGATGCTCCAGTTCCATCTCTGTCTCCCACAGTCGAGCCCCGTTGAGGATGGAGCCCACCTCAGGCTTTGCAGAGCAACCTG ......................................................................................((((.((((..........(((((((..(((.((...)).)))...)))))))))))....))))..((((.(((...........))).))))...................................................................... ............................................................................77...........................................................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................TGAAGAGCCCGGCACGATTATTCCTTA................................................................................................................................................................................................. | 27 | 1 | 19.00 | 19.00 | 5.00 | 5.00 | 2.00 | - | - | - | - | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................TGGACTCCTCATTCCTGGGCCTGGCGG.................................................................................................................................. | 27 | 1 | 9.00 | 9.00 | - | 1.00 | 1.00 | - | 2.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TACCTGGCGCTCTGGACTCCTCATTCC.............................................................................................................................................. | 27 | 1 | 7.00 | 7.00 | - | - | - | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................TACCTGGCGCTCTGGACTCCTCATTC............................................................................................................................................... | 26 | 1 | 6.00 | 6.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGAAGCTGACGGATGATGCTCCAGTTC.................................................................... | 27 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGAAGCTGACGGATGATGCTCCAGTT..................................................................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGAAGAGCCCGGCACGATTATTCCTT.................................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGAAGCTGACGGATGATGCTCCAGT...................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CCTGGCGCTCTGGACTCCTCATT................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGATGCTCCAGTTCCATCTCTGTCTCCa...................................................... | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........ACGTGGCCTCGTCAGAGCCCTGAAGA...................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TCGAGCCCCGTTGAGGATGGAGCCCA........................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGACTCCTCATTCCTGGGCCTGGCGGG................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CCTCGTCAGAGCCCTGAAGAGCCCG................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............TGGCCTCGTCAGAGCCtga.......................................................................................................................................................................................................................... | 19 | tga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................AGCTGACGGATGATGCTCCAGT...................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................TACCTGGCGCTCTGGACTCCTCATTCCTG............................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................CCCACTGAAGCTGACGGATGATGCTCC......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGAAGAGCCCGGCACGATTATTCCT................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGAAGCTGACGGATGATGCTCCAGTTa.................................................................... | 27 | a | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGACTCCTCATTCCTGGGCCTGGC.................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TACCTGGCGCTCTGGACTCCTCATT................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CACAGTCGAGCCCCGTTGAGGATGca............................. | 26 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................CACGATTATTCCTTATGCGGCCGGGG...................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGACTCCTCATTCCTGGGCCTGGCGGGA................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TCCCAGGACAGACCCCCGCCCCACTGA............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................GAAGCTGACGGATGATGCTCCAGT...................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGACGGATGATGCTCCAGTTCCATCTC.............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TCCCACAGTCGAGCCCCGTTGAGGATGGA............................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGTCGAGCCCCGTTGAGGATGGAG............................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTTGGGACCACGTGGCCTCGTCAGAGCCCTGAAGAGCCCGGCACGATTATTCCTTATGCGGCCGGGGAATAAGCCAACTCTACCTGGCGCTCTGGACTCCTCATTCCTGGGCCTGGCGGGACCCACCACATCCCAGGACAGACCCCCGCCCCACTGAAGCTGACGGATGATGCTCCAGTTCCATCTCTGTCTCCCACAGTCGAGCCCCGTTGAGGATGGAGCCCACCTCAGGCTTTGCAGAGCAACCTG ......................................................................................((((.((((..........(((((((..(((.((...)).)))...)))))))))))....))))..((((.(((...........))).))))...................................................................... ............................................................................77...........................................................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................GTTCCATCTCTGTCTCCCACAGTCGA.............................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGATGATGCTCCAGTTCCATCTCTGTC.......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................GTCTCCCACAGTCGAGCCCCGTTGA.................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................ATCTCTGTCTCCCACAGTCGAG............................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GCTGACGGATGATGCTCCAGTTCCA.................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |