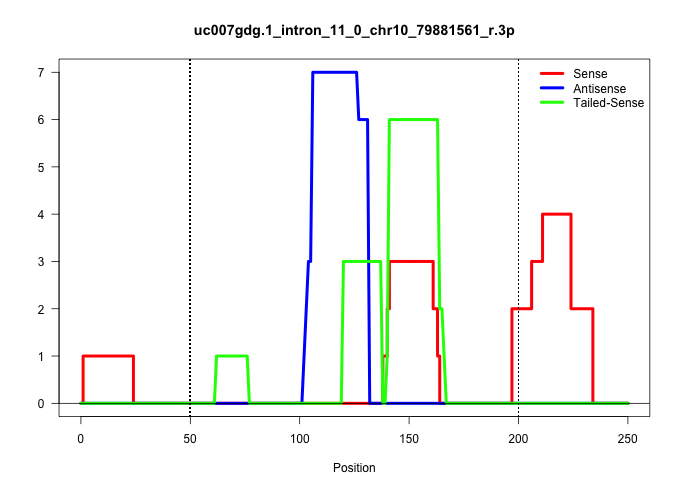
| Gene: Tcfe2a | ID: uc007gdg.1_intron_11_0_chr10_79881561_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) TDRD1.ip |
(23) TESTES |
| CTTGGAACTCACAGAGATTGCCTGCCTCTGTCTCCTCTATGCTGGCAGAGCACAGGTTTTTAGTGGCTGCATTAACAAGGCCCCTCAAGTGTCAGGGATGTGGCATCCTTGGAACCTGACTACAGCATGGTAGGGGGCCACATGAGGGCATGTGGAGATGGCAGTGGCCAGACCTTCCTCTCTGCTCTTGCTCCCAGCAGATACTCAGCCGAAGAAGGTCCGGAAGGTTCCGCCTGGTCTCCCTTCCTCG .........................................................................................................((....))..(((.((..((((.((((((((((((((((....)))))))...((((....)))).......)))))))))..))))...))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | mjTestesWT4() Testes Data. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAaa..................................................................................... | 24 | aa | 7.00 | 4.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCA....................................................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CATGAGGGCATGTGGAGATGGCAa...................................................................................... | 24 | a | 3.00 | 0.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAa...................................................................................... | 23 | a | 3.00 | 4.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGATACTCAGCCGAAGAAGGTCCGGA.......................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TACAGCATGGTAGGagac................................................................................................................ | 18 | agac | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGAGGGCATGTGGAGATGGCAGTGG................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAt...................................................................................... | 23 | t | 2.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAG...................................................................................... | 23 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CATGAGGGCATGTGGAGATGGCAt...................................................................................... | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAtga.................................................................................... | 25 | tga | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AGCCGAAGAAGGTCCGGAAGGTTCCGCC................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................TACAGCATGGTAGGtgac................................................................................................................ | 18 | tgac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CATGAGGGCATGTGGAGATGG......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAtag.................................................................................... | 25 | tag | 1.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CACATGAGGGCATGTGGAGATGGCAG...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TTGGAACTCACAGAGATTGCCTG.................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AAGAAGGTCCGGAAGGTTCCGCC................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGGCAtttt................................................................................... | 26 | tttt | 1.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GTGGCATCCTTGGAACCTGACTAC............................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GTGGCTGCATTAACg............................................................................................................................................................................. | 15 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................ATGAGGGCATGTGGAGATGacag...................................................................................... | 23 | acag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTGGAACTCACAGAGATTGCCTGCCTCTGTCTCCTCTATGCTGGCAGAGCACAGGTTTTTAGTGGCTGCATTAACAAGGCCCCTCAAGTGTCAGGGATGTGGCATCCTTGGAACCTGACTACAGCATGGTAGGGGGCCACATGAGGGCATGTGGAGATGGCAGTGGCCAGACCTTCCTCTCTGCTCTTGCTCCCAGCAGATACTCAGCCGAAGAAGGTCCGGAAGGTTCCGCCTGGTCTCCCTTCCTCG .........................................................................................................((....))..(((.((..((((.((((((((((((((((....)))))))...((((....)))).......)))))))))..))))...))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | mjTestesWT4() Testes Data. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................CCTTGGAACCTGACTACAGCATGGTA...................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................TCCTCTCTGCTCTTGCTCCCAGCAGAt................................................. | 27 | t | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 |
| ........................................................................................................ATCCTTGGAACCTGACTACAGCATGGTA...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................GAGCACAGGTTTTTAata............................................................................................................................................................................................ | 18 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TCCTCTATGCTGGCAGAGCACAGGTTTTTAt............................................................................................................................................................................................ | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................CCTTGGAACCTGACTACAGCATGGTAt...................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GCATCCTTGGAACCTGACTACAGCATGGTA...................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CATCCTTGGAACCTGACTACAGCA........................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |