
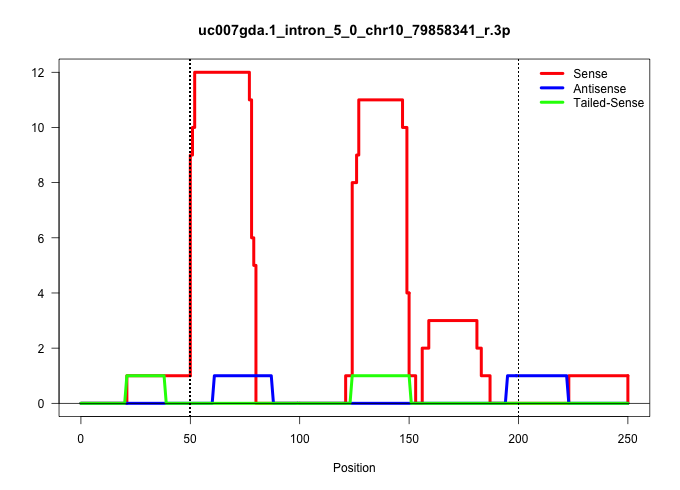
| Gene: Mbd3 | ID: uc007gda.1_intron_5_0_chr10_79858341_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(14) TESTES |
| CCCGAGAGGTACAGGCCCCCATACCTCATAGGGCCTCCTTAGGCTTCCCAGATGAACTGTAGCCACCTAGCTCAAGGCTTGACTGCTAGGGCTGCTCTTCCCACGTGCCTTCCTAATGACCTGGTGAAAACAGGCATGGAGAACAGTGGTGGTGGGTAGGAGATGGGTCCTGCTCGGGCTGACCAGCCTCCCGTCCGCAGCCCCAGCGGGAAGAAGTTCCGCAGCAAGCCACAACTGGCACGTTACCTGG .....................................................(((..(((((..(((((((((.....)))..)))))))))))..)))((((.((..((((..(((.((((.......)))))))))))..))))))..................................................................................................... ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GATGAACTGTAGCCACCTAGCTCAAGGC............................................................................................................................................................................ | 28 | 1 | 8.00 | 8.00 | - | - | 1.00 | 2.00 | - | - | 1.00 | 1.00 | 2.00 | 1.00 | - | - | - | - |
| ...............................................................................TGACTGCTAGGGCTGCTCTTCCCACG................................................................................................................................................. | 26 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGAAAACAGGCATGGAGAACAGTGG..................................................................................................... | 25 | 1 | 4.00 | 4.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................TGAAAACAGGCATGGAGAACAGTGGT.................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGAACTGTAGCCACCTAGCTCAAGGCTT.......................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................AAACAGGCATGGAGAACAGTGG..................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GATGAACTGTAGCCACCTAGCTCAAGGCTT.......................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAGGAGATGGGTCCTGCTCGGGCTG..................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GATGAACTGTAGCCACCTAGCTCAAGG............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GATGAACTGTAGCCACCTAGCTCAAGGCT........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................TACCTCATAGGGCCTCCTTAGGCTTCCCA........................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................ATGAACTGTAGCCACCTAGCTCAAGGCTT.......................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................TACCTCATAGGGCCTtta................................................................................................................................................................................................................... | 18 | tta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................TGAAAACAGGCATGGAGAACAGTGGTt................................................................................................... | 27 | t | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................AAAACAGGCATGGAGAACAGTGGTGGT................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GCAAGCCACAACTGGCACGTTACCTGG | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................TAGGAGATGGGTCCTGCTCGGGCTGAC................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGGTGAAAACAGGCATGGAGAACAGT....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GAGATGGGTCCTGCTCGGGCTGACCAGC............................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................TGACTGCTAGGGCTGCTCTTCCCACa................................................................................................................................................. | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| CCCGAGAGGTACAGGCCCCCATACCTCATAGGGCCTCCTTAGGCTTCCCAGATGAACTGTAGCCACCTAGCTCAAGGCTTGACTGCTAGGGCTGCTCTTCCCACGTGCCTTCCTAATGACCTGGTGAAAACAGGCATGGAGAACAGTGGTGGTGGGTAGGAGATGGGTCCTGCTCGGGCTGACCAGCCTCCCGTCCGCAGCCCCAGCGGGAAGAAGTTCCGCAGCAAGCCACAACTGGCACGTTACCTGG .....................................................(((..(((((..(((((((((.....)))..)))))))))))..)))((((.((..((((..(((.((((.......)))))))))))..))))))..................................................................................................... ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................GCCCCAGCGGGAAGAAGTTCCGCAGCAa........................ | 28 | a | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGCTCTTCCCACGTGCCTTCCTAAta...................................................................................................................................... | 26 | ta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CGCAGCCCCAGCGGGAAGAAGTTCCGCA........................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GGTGGTGGGTAGGAgttc......................................................................................... | 18 | gttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................GCCACCTAGCTCAAGGCTTGACTGCTA.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |