
| Gene: Ppap2c | ID: uc007fyr.1_intron_4_0_chr10_78993774_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.mut |
(11) TESTES |
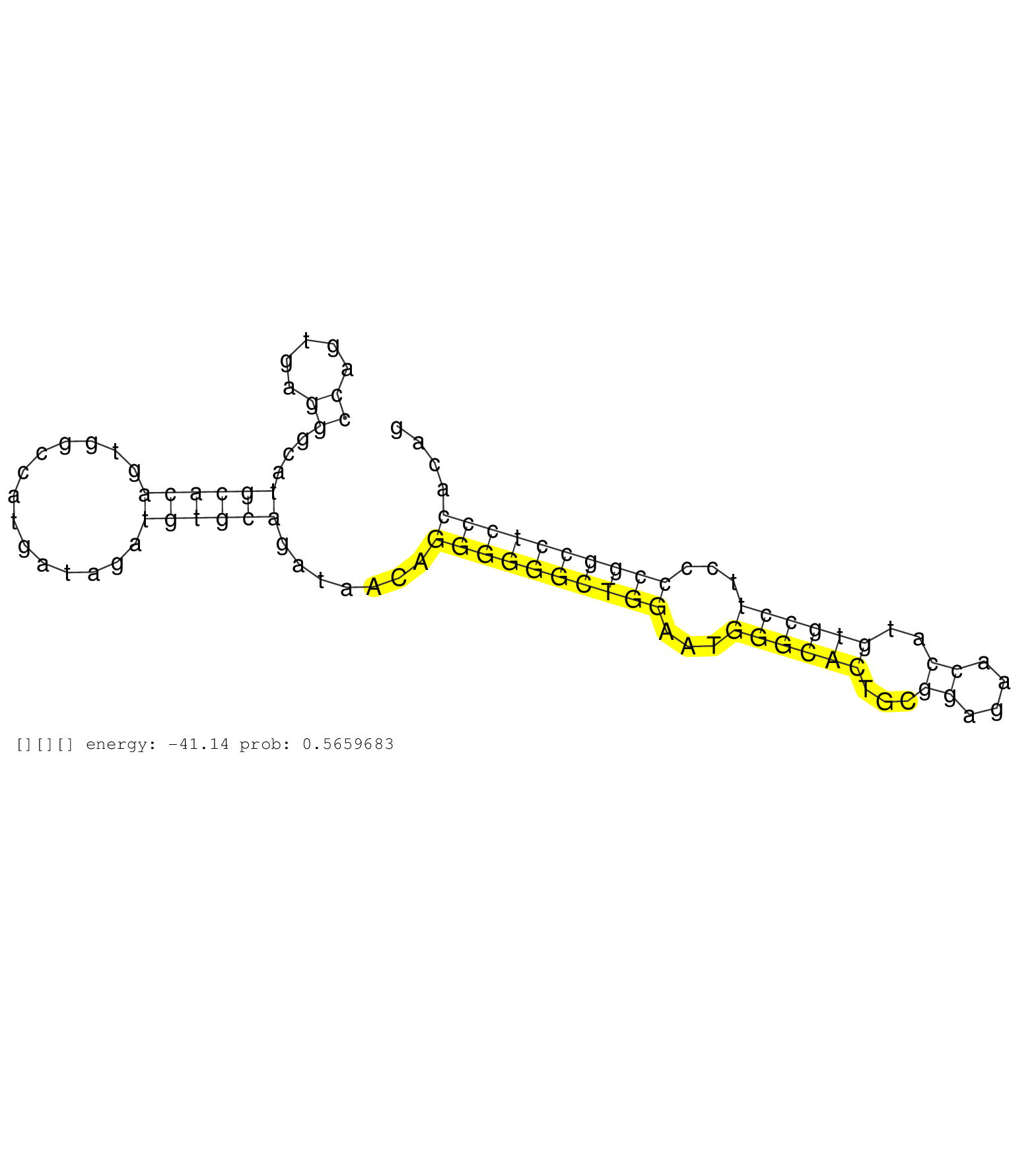
| TGAAGAGGCAGCTCTAGGGAGCCCTGAAGTGAGGAGAGGCTATAGCTGGGTAGAGCTGCTGAGTAGTTGGGCTCATTCAAAGAAGCCAATGGAGTAGTTTCCAGTGAGGGCATGCACAGTGGCCATGATAGATGTGCAGATAACAGGGGGGCTGGAATGGGCACTGCGGAGAACCATGTGCCTTCCCCGGCCTCCCACAGCCTCTCTGCCTTTCATCATCCTGACCCTGGTGAATGCACCATATAAGCGG ....................................................................................................((.....))...((((((..............)))))).......((((((((((...((((((...((....))..))))))...))))))))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................AACAGGGGGGCTGGAATGGGCAtat.................................................................................... | 25 | tat | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TCCTGACCCTGGTGAATGCA............ | 20 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGGGGGGCTGGAATGGGCA....................................................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGCCTTTCATCATCCTGACCCTGGTGA................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................ACAGGGGGGCTGGAATGGGCACTaa................................................................................... | 25 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................CAGGGGGGCTGGAATGGGCACTat................................................................................... | 24 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................TCCCACAGCCTCTCgatg........................................ | 18 | gatg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................CAGGGGGGCTGGAATGGGCACTt.................................................................................... | 23 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................ACAGGGGGGCTGGAATGGGCAC...................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGGGGGGCTGGAATGGGCACTGaga................................................................................. | 25 | aga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGGGGGGCTGGAATGGGCAaga.................................................................................... | 23 | aga | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| TGAAGAGGCAGCTCTAGGGAGCCCTGAAGTGAGGAGAGGCTATAGCTGGGTAGAGCTGCTGAGTAGTTGGGCTCATTCAAAGAAGCCAATGGAGTAGTTTCCAGTGAGGGCATGCACAGTGGCCATGATAGATGTGCAGATAACAGGGGGGCTGGAATGGGCACTGCGGAGAACCATGTGCCTTCCCCGGCCTCCCACAGCCTCTCTGCCTTTCATCATCCTGACCCTGGTGAATGCACCATATAAGCGG ....................................................................................................((.....))...((((((..............)))))).......((((((((((...((((((...((....))..))))))...))))))))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................GCCTCCCACAGCCTCTCT........................................... | 18 | 3 | 1.67 | 1.67 | - | - | - | - | 1.33 | - | - | - | - | - | 0.33 |