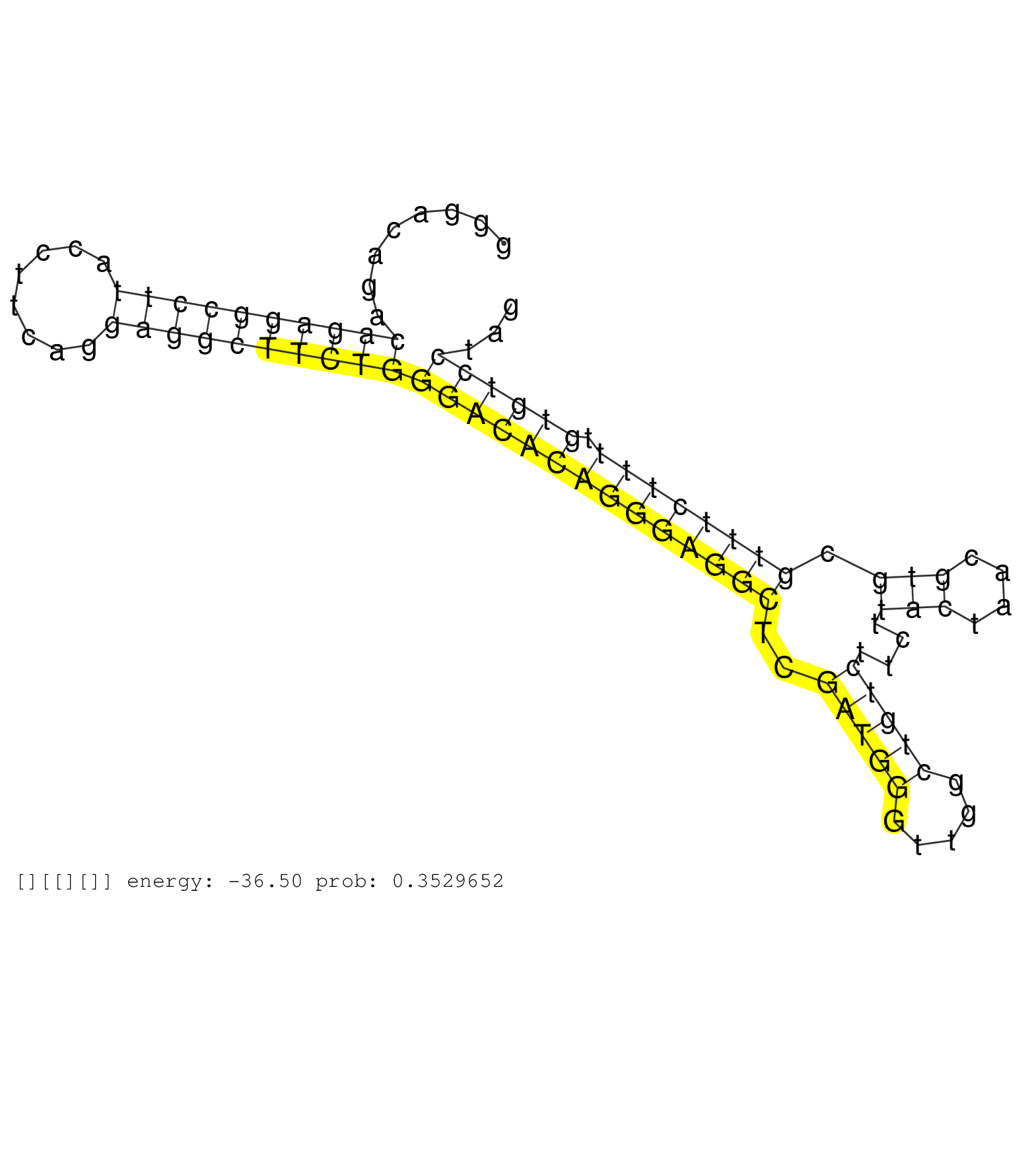
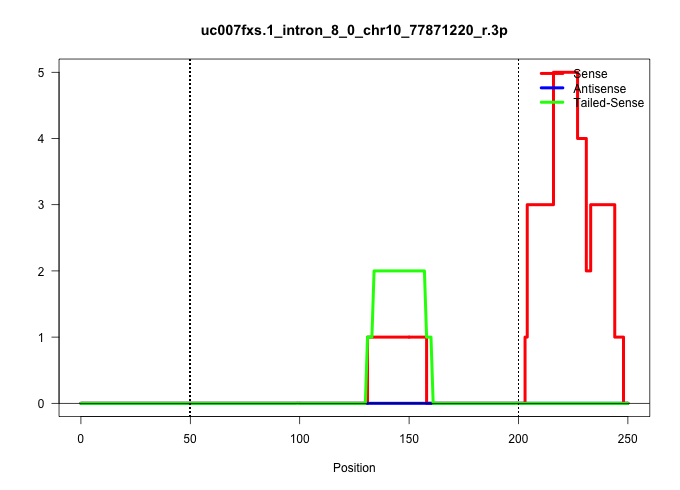
| Gene: Rrp1 | ID: uc007fxs.1_intron_8_0_chr10_77871220_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CAGCGCCTGCCCTGACCATTCCAATCAGCTCTAATCTGTAGTTTCTGCTCACTCAGCCAAGTACAGAGCTTTGATCTCCAAGGCCTCAATCACGCCCCGTGGGACAGACAGAGGCCTTACCTTCAGGAGGCTTCTGGGACACAGGGAGGCTCGATGGGTTGGCTGTCTTCTTACTAACGTGCGTTTCTTTTGTGTCCTAGCTCATGAGGATGGTCCTAAGTGAGTCACTGAAGGCTGTGAAAGCCCGGGG ............................................................................................................((((((((((........))))))))))((((((((((((((..(((((.....)))))....(((....))).)))))))).))))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................TAAGTGAGTCACTGAAGGCTG............. | 21 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................AACGTGCGTTTCTTTTGTGTCCT.................................................... | 23 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAGGATGGTCCTAAGTGAGTCACTGA................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 |
| ........................................................................................................................................................................................................................TAAGTGAGTCACTGAAGGCTGTGAAAGC...... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TTCTGGGACACAGGGAGGCTCGATGGt............................................................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................TTCTGGGACACAGGGAGGCTCGATGGG............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGACACAGGGAGGCTCGATGGGTTt......................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................CTGTGAAAGCCCGGG. | 15 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................GCTGTGAAAGCCCGG.. | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................................GCTGTGAAAGCCCGGG. | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TTCTGGGACACAGGGAGGCTCG................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................TCTGGGACACAGGGAGGCTCGATGGGTTGGt....................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................ATGAGGATGGTCCTAAGTGAGTCA....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| CAGCGCCTGCCCTGACCATTCCAATCAGCTCTAATCTGTAGTTTCTGCTCACTCAGCCAAGTACAGAGCTTTGATCTCCAAGGCCTCAATCACGCCCCGTGGGACAGACAGAGGCCTTACCTTCAGGAGGCTTCTGGGACACAGGGAGGCTCGATGGGTTGGCTGTCTTCTTACTAACGTGCGTTTCTTTTGTGTCCTAGCTCATGAGGATGGTCCTAAGTGAGTCACTGAAGGCTGTGAAAGCCCGGGG ............................................................................................................((((((((((........))))))))))((((((((((((((..(((((.....)))))....(((....))).)))))))).))))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................GCTTTGATCTCCAAGaat........................................................................................................................................................................ | 18 | aat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....CCCTGACCATTCCAacgc................................................................................................................................................................................................................................... | 18 | acgc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |