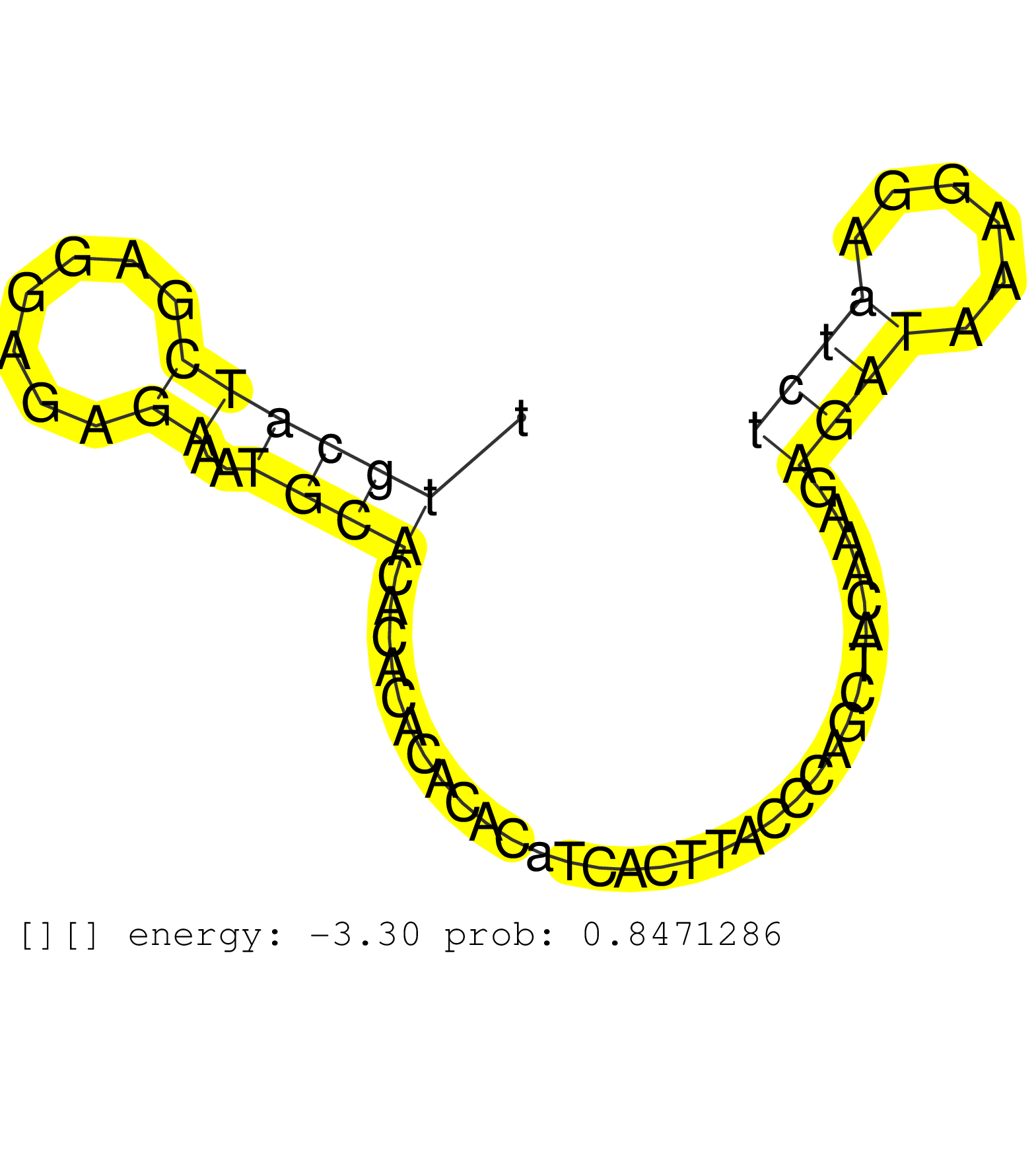
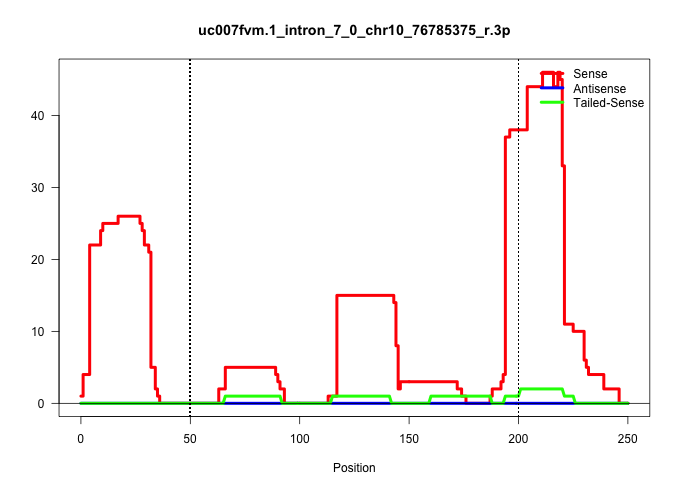
| Gene: Adarb1 | ID: uc007fvm.1_intron_7_0_chr10_76785375_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| TTGATAAACTAAGTAGAAGAGGCAATAGGTCCATTAATTAAAAAAAAAAAGTTTCAAAGTGCATTCTAGCCCCGAGCAGTTACATCCTTGGTCAATGTATAGTTCCTCACGTTTGCATCGAGGAGAGAAATGCACACACACACACATCACTTACCCAGCTACAAAGAGATAAAGGAATCTATAAATTTTGCATTTACAAGATCCTGCAACGAAGGCGTTGTAAGTTACTCTTTCTGGGCACCACAGGTTC .................................................................................................................((((((.......))..))))................................((((......))))...................................................................... ................................................................................................................113................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................TACAAGATCCTGCAACGAAGGCGTTGT............................. | 27 | 1 | 22.00 | 22.00 | 7.00 | 6.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ....TAAACTAAGTAGAAGAGGCAATAGGTCC.......................................................................................................................................................................................................................... | 28 | 1 | 16.00 | 16.00 | 4.00 | 8.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TACAAGATCCTGCAACGAAGGCGTTG.............................. | 26 | 1 | 11.00 | 11.00 | 3.00 | 3.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................TCGAGGAGAGAAATGCACACACACACAC......................................................................................................... | 28 | 1 | 6.00 | 6.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCGAGGAGAGAAATGCACACACACACA.......................................................................................................... | 27 | 1 | 6.00 | 6.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGCAACGAAGGCGTTGTAAGTTACTC.................... | 26 | 1 | 4.00 | 4.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................AAGGCGTTGTAAGTTACTCTTTCTGGGC........... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................TGTAAGTTACTCTTTCTGGGCACCACAG.... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAGCCCCGAGCAGTTACATCCTTGGTC............................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| .TGATAAACTAAGTAGAAGAGGCAATAGG............................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCGAGGAGAGAAATGCACACACACACACA........................................................................................................ | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCACTTACCCAGCTACAAAGAGATAAAGGA.......................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGCATCGAGGAGAGAAATGCACACACACAC........................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TAAGTAGAAGAGGCAATAGGTCCATTA...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGCAACGAAGGCGTTGTAAGTTACTCTT.................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAGCCCCGAGCAGTTACATCCTTGG............................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TTACAAGATCCTGCAACGAAGGCGTTG.............................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TAAGTAGAAGAGGCAATAGGTCCATT....................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCACTTACCCAGCTACAAAGAGATAA.............................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TACAAGATCCTGCAACGAAGGCGTTat............................. | 27 | at | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGATAAACTAAGTAGAAGAGGCAATAG.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TTCTAGCCCCGAGCAGTTACATCCTTG................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................ACAAAGAGATAAAGGAATCTATAAATgt.............................................................. | 28 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................TTGCATTTACAAGATCCTGCAACGAAGGC.................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGCAACGAAGGCGTTGTAAGTTACTCT................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AAGTAGAAGAGGCAATAGGTCCAT........................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................AGAGGCAATAGGTCCAT........................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CATCGAGGAGAGAAATGCACACACctc............................................................................................................ | 27 | ctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................TCACTTACCCAGCTACAAAGAGATAAAG............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TCCTGCAACGAAGGCGTTGTAggtt........................ | 25 | ggtt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTTACAAGATCCTGCAACGAAGGCGTT............................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TTCTAGCCCCGAGCAGTTACATCCTT................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....TAAACTAAGTAGAAGAGGCAATAGGTCCAT........................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAGCCCCGAGCAGTTACATCCTTGGa.............................................................................................................................................................. | 26 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CAAGATCCTGCAACGAAGGCGTTGTAAGT......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....TAAACTAAGTAGAAGAGGCAATAGGTC........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TGCATTTACAAGATCCTGCAACGAAGGC.................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTGATAAACTAAGTAGAAGAGGCAATAGGTCCATTAATTAAAAAAAAAAAGTTTCAAAGTGCATTCTAGCCCCGAGCAGTTACATCCTTGGTCAATGTATAGTTCCTCACGTTTGCATCGAGGAGAGAAATGCACACACACACACATCACTTACCCAGCTACAAAGAGATAAAGGAATCTATAAATTTTGCATTTACAAGATCCTGCAACGAAGGCGTTGTAAGTTACTCTTTCTGGGCACCACAGGTTC .................................................................................................................((((((.......))..))))................................((((......))))...................................................................... ................................................................................................................113................................................................180.................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|