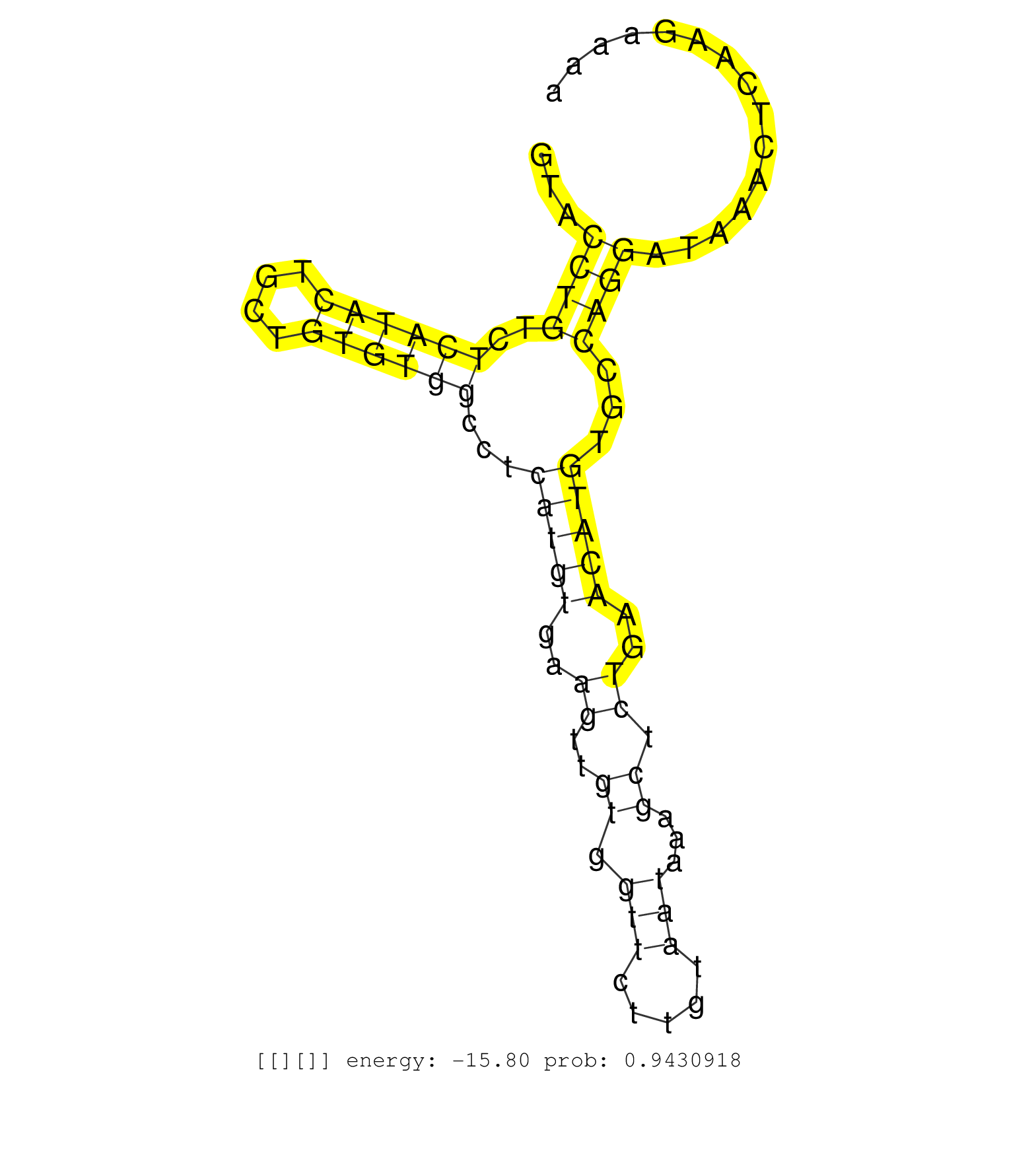

| Gene: Adarb1 | ID: uc007fvl.1_intron_7_0_chr10_76785329_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| CTCATTATCATCATGGGACCATCATTTTTATTCCAGAAACATGCCTCTAGGTACCTGTCTCATACTGCTGTGTGGCCTCATGTGAAGTTGTGGTTCTTGTAATAAAGCTCTGAACATGTGCCAGGATAAACTCAAGAAAACCTTTTTTGATTGACTCACATTCCTGGTCTCTGGGATAACAGTTCGTAAAGCTCCCTTCAGACACAGTCGCTGCTCCTGCCTCGGAGGGAAGGGAGTCCGCATGATCTTT .....................................................((((..((((((....))))))...(((((..((..((.(((.....)))...)).))..)))))...))))............................................................................................................................. ..................................................51.......................................................................................140............................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................TGAACATGTGCCAGGATAAACTCAAG.................................................................................................................. | 26 | 1 | 7.00 | 7.00 | - | 2.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................TAATAAAGCTCTGAACATGTGCCAGG............................................................................................................................. | 26 | 1 | 6.00 | 6.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAACATGTGCCAGGATAAACTCAAGA................................................................................................................. | 27 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGTGAAGTTGTGGTTCTTGTAATAAAGC.............................................................................................................................................. | 28 | 1 | 5.00 | 5.00 | - | - | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTACCTGTCTCATACTGCTGTGT................................................................................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................TAGGTACCTGTCTCATACTGCTGTGTGGC.............................................................................................................................................................................. | 29 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TCGTAAAGCTCCCTTCAGACACAGTCGC....................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCCTCGGAGGGAAGGGAGTCCGCATG...... | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAACAGTTCGTAAAGCTCCCTTCAG................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................TGTGGTTCTTGTAATAAAGCTCTGAAC....................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .TCATTATCATCATGGGACCATCATTT............................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ......................................................................................................TAAAGCTCTGAACATGTGCCAGGATA.......................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............TGGGACCATCATTTTTATTCCAGAAACATGC.............................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................TGTGCCAGGATAAACTCAAGAAAACCTTT......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTACCTGTCTCATACTGCTGc................................................................................................................................................................................... | 24 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGGTCTCTGGGATAACAGTTCGTAAA............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................CTCTGAACATGTGCCAGGA............................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................................TGCCTCGGAGGGAAGGGAGTCCGCAT....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGAAGTTGTGGTTCTTGTAATAAAGCT............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTGTAATAAAGCTCTGAACATGTGCCA............................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTTATTCCAGAAACATGCCTCTAGGTA..................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTACCTGTCTCATACTGCTGTG.................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCATACTGCTGTGTGGCCTCATGTG...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CACAGTCGCTGCTCCTGCCTCGGA........................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCCTCGGAGGGAAGGGAGTCCGCct....... | 26 | ct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTTTTATTCCAGAAACATGCCTCTAGG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TACTGCTGTGTGGCCTCATGTGAAG................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CTTGTAATAAAGCTCTGAACATGTGCCA............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAACATGTGCCAGGATAAACTCAAGAAA............................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................ATAAAGCTCTGAACATGTGCCAGGATA.......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGAAGTTGTGGTTCTTGTAAT................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTACCTGTCTCATACTG....................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGGTTCTTGTAATAAAGCTCTGAACA...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................TCGGAGGGAAGGGAGTCCGCATGATa... | 26 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TCTCTGGGATAACAGTTCGTAAAGCTCC....................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CAGTCGCTGCTCCTGCCTCGGAGGGta................... | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................TGCTGTGTGGCCTCATGTGAAGTTGT............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGTGCCAGGATAAACTCAAGAAAACCTTTT........................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ACATGTGCCAGGATAAACTCAAGAAAACC............................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GTCGCTGCTCCTGCCTCGGAGGGAAGGG................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCGCTGCTCCTGCCTCGGAGGGAAG.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAACAGTTCGTAAAGCTCCCTTCAGAC............................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| CTCATTATCATCATGGGACCATCATTTTTATTCCAGAAACATGCCTCTAGGTACCTGTCTCATACTGCTGTGTGGCCTCATGTGAAGTTGTGGTTCTTGTAATAAAGCTCTGAACATGTGCCAGGATAAACTCAAGAAAACCTTTTTTGATTGACTCACATTCCTGGTCTCTGGGATAACAGTTCGTAAAGCTCCCTTCAGACACAGTCGCTGCTCCTGCCTCGGAGGGAAGGGAGTCCGCATGATCTTT .....................................................((((..((((((....))))))...(((((..((..((.(((.....)))...)).))..)))))...))))............................................................................................................................. ..................................................51.......................................................................................140............................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|