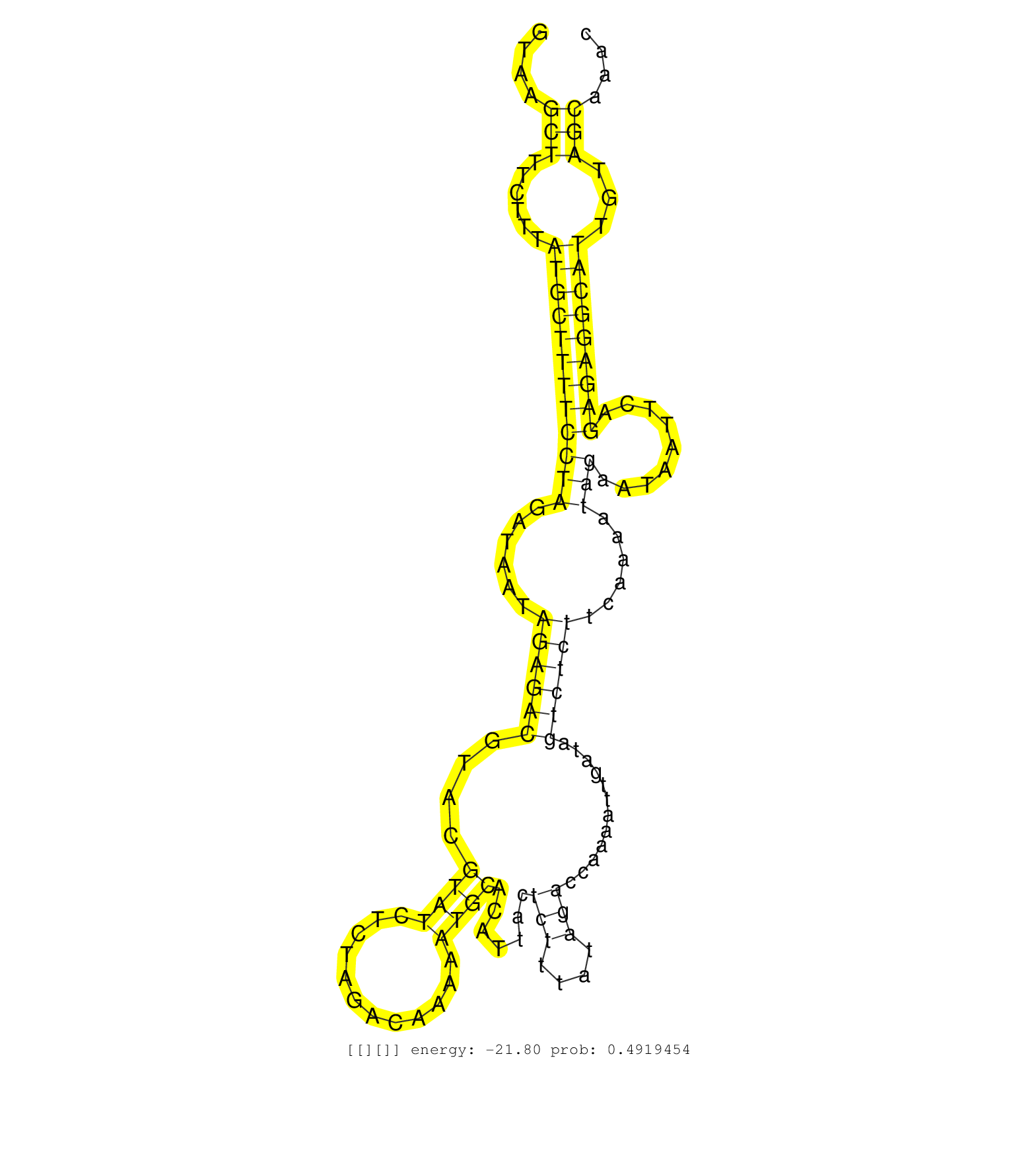

| Gene: Ddx50 | ID: uc007fhq.1_intron_1_0_chr10_62090431_r.5p | SPECIES: mm9 |
 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| AAACTAAGAAGAATGTTACTGAAATGGCCATGAATCCACACATAAAGCAGGTAAGCTTTCTTTATGCTTTTCCTAGATAATAGAGACGTACGTATCTCTAGACAAAAATGCACATTACTCTTTATAGACCAAAATTGATAGTCTCTTCAAAATAGAATAATTCAGAGAGGCATTGTAGCAAACTTTTTCTCATATATTAAGTCATAATAGGTATTTTTGTTTATTTGTTTTTTCCCTTCTGCTAACAATC ......................................................(((......((((((((((((......((((((....((((............)))).......(((....)))............))))))......))).........)))))))))...)))....................................................................... ..................................................51..................................................................................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGCTTTCTTTATGCTTTTCCTAGATAATAGAGACGTACGTATCTCTAGA.................................................................................................................................................... | 52 | 1 | 70.00 | 70.00 | 70.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................ACACATAAAGCAGGTAAGC.................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TTTTTTCCCTTCTGCagaa.... | 19 | agaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................TACTGAAATGGCCATGAATCCACACATAta............................................................................................................................................................................................................ | 30 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................AATGGCCATGAATCCACACATAAAGCAGG....................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................ATAATTCAGAGAGGCATTGTAGC....................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................TCCTAGATAATAGAGAga.................................................................................................................................................................. | 18 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....TAAGAAGAATGTTACTGAAATGGCCATG.......................................................................................................................................................................................................................... | 28 | 2 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....TAAGAAGAATGTTACTGAAATGGCCA............................................................................................................................................................................................................................ | 26 | 2 | 1.00 | 1.00 | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ...CTAAGAAGAATGTTACTGAAATGGCC............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TAGAATAATTCAGAGAGGCATTGTAGC....................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CTCTTTATAGACCAAAca................................................................................................................... | 18 | ca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........AGAATGTTACTGAAATGGCCATGAATCC..................................................................................................................................................................................................................... | 28 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ......AGAAGAATGTTACTGAAATGGCCATG.......................................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........AATGTTACTGAAATGGCCATGAATCC..................................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| .........AGAATGTTACTGAAATGGCCATGAATC...................................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ................TACTGAAATGGCCATGAATCC..................................................................................................................................................................................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...................TGAAATGGCCATGAATCCACAC................................................................................................................................................................................................................. | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .....AAGAAGAATGTTACTGAAATGGCCATGAATCC..................................................................................................................................................................................................................... | 32 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| AAACTAAGAAGAATGTTACTGAAATGGCCATGAATCCACACATAAAGCAGGTAAGCTTTCTTTATGCTTTTCCTAGATAATAGAGACGTACGTATCTCTAGACAAAAATGCACATTACTCTTTATAGACCAAAATTGATAGTCTCTTCAAAATAGAATAATTCAGAGAGGCATTGTAGCAAACTTTTTCTCATATATTAAGTCATAATAGGTATTTTTGTTTATTTGTTTTTTCCCTTCTGCTAACAATC ......................................................(((......((((((((((((......((((((....((((............)))).......(((....)))............))))))......))).........)))))))))...)))....................................................................... ..................................................51..................................................................................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|