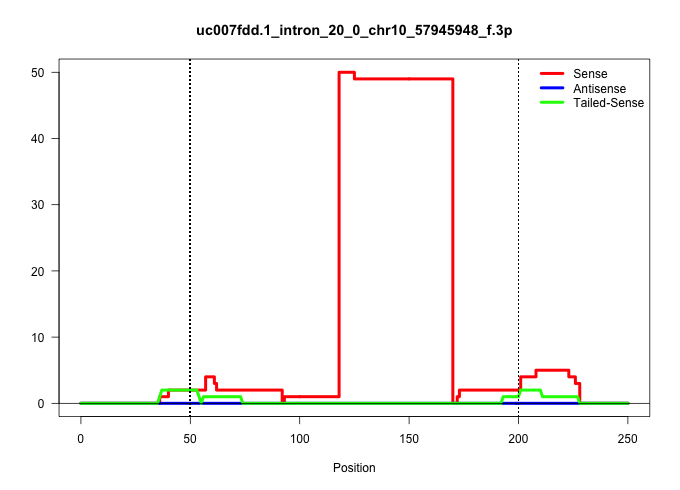
| Gene: Ranbp2 | ID: uc007fdd.1_intron_20_0_chr10_57945948_f.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| GTGTTGAAATGACAGAATGGCTAAGCATGAGGGGAGCACAAATGGGAACTATGAACAGCAGTCGGAGGTTAAAGGCCAGATTGTCAGAGCTCAGGGCCATGTGTGAGAGAGGGAGAGTGTGAAGATCATTTCTAGGTTTGTGGATGGGAAAACCTACCTACTAAAATTATTTTGCTCAATATGATCAATTGCATTTGAAGATGAAGATTATGAAATGGCTGTCAAGAAGCTTAATGGAAAACTCTATCTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................GTGAAGATCATTTCTAGGTTTGTGGATGGGAAAACCTACCTACTAAAATTAT................................................................................ | 52 | 1 | 49.00 | 49.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAAGATTATGAAATGGCTGTCAAGAA...................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GCAGTCGGAGGTTAAAGGCCAGATTGTCAGAGCTC.............................................................................................................................................................. | 35 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGCTCAATATGATCAATTGCATTTGAAG.................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................TTTGAAGATGAAGActtc....................................... | 18 | cttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................ATGAAGATTATGAAATGGCTGTC........................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................ACAAATGGGAACTATGct................................................................................................................................................................................................... | 18 | ct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ATGAAGATTATGAAATGGCTGTCAAG........................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................GGGCCATGTGTGAGAGAGGGAGAGTGTGAAGA............................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................AGCAGTCGGAGGTTgctt................................................................................................................................................................................ | 18 | gctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................AATGGGAACTATGAACAGCAGT............................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................CACAAATGGGAACTATGAACAGCAG............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................GCTCAATATGATCAATTGCATTTGAAG.................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TATGAAATGGCTGTCAAGAA...................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CACAAATGGGAACTATGt.................................................................................................................................................................................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAAGATTATGAAATGGCTGTCAAGAt...................... | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| GTGTTGAAATGACAGAATGGCTAAGCATGAGGGGAGCACAAATGGGAACTATGAACAGCAGTCGGAGGTTAAAGGCCAGATTGTCAGAGCTCAGGGCCATGTGTGAGAGAGGGAGAGTGTGAAGATCATTTCTAGGTTTGTGGATGGGAAAACCTACCTACTAAAATTATTTTGCTCAATATGATCAATTGCATTTGAAGATGAAGATTATGAAATGGCTGTCAAGAAGCTTAATGGAAAACTCTATCTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................CACAAATGGGAACTATaa...................................................................................................................................................................................................... | 18 | aa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |