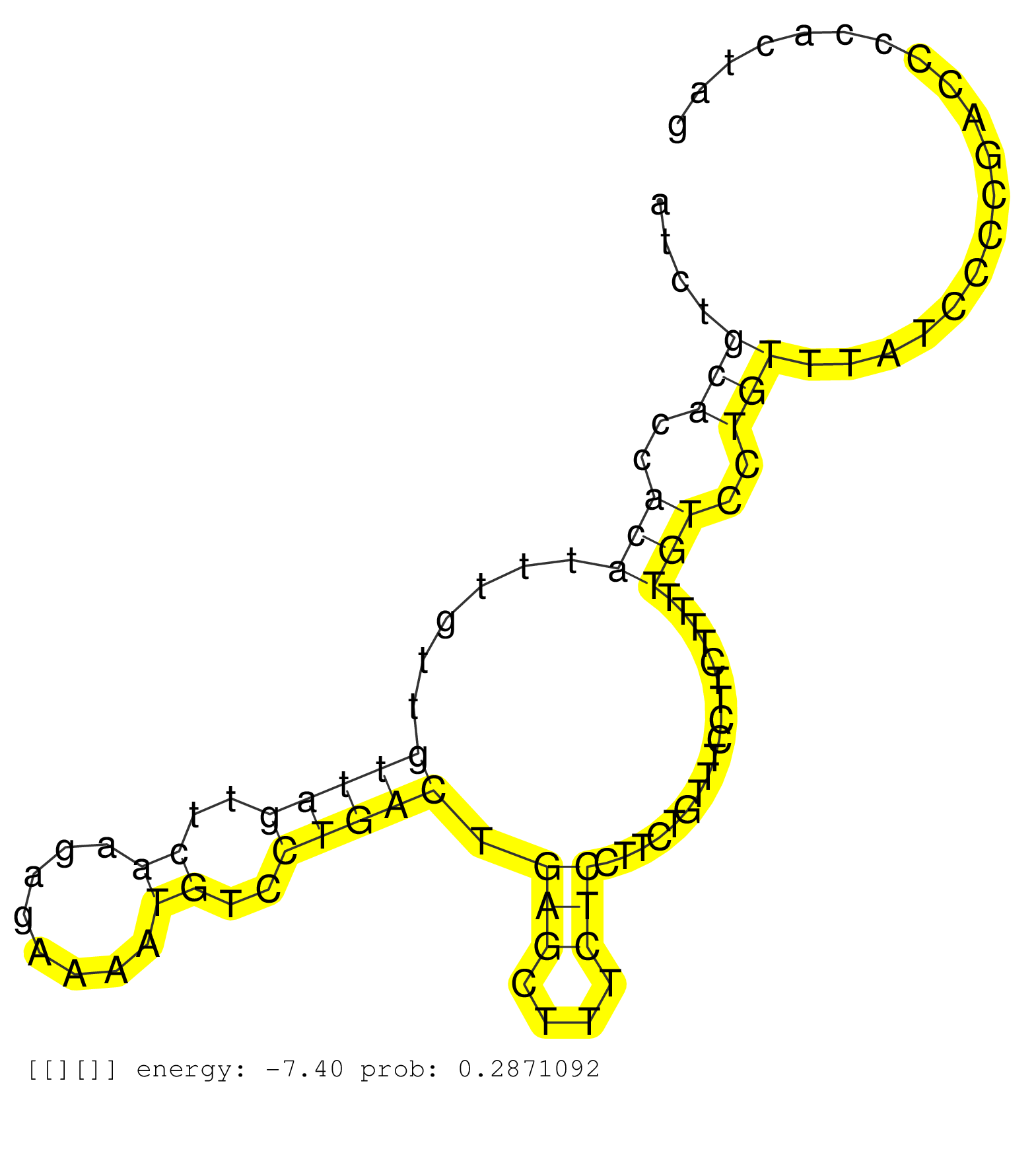
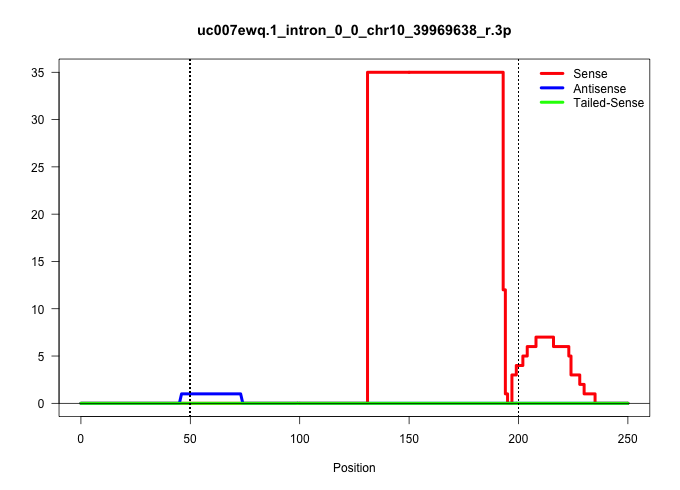
| Gene: Gtf3c6 | ID: uc007ewq.1_intron_0_0_chr10_39969638_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(1) PIWI.mut |
(9) TESTES |
| ACAGCTGGCTCGGACAGTGCTACCCTTTTTCATAGGGCACCTTCACGCTGACGTCTGAGTGGTTGTACAATTCACATCCCCACTGGCGGTGTAGGCTCTCATCTGCACCACATTTGTTGTTAGTTCAAGAGAAAATGTCCTGACTGAGCTTTCTCCTTCTGTTTCCTTCTTTTTGTCCTGTTTATCCCCGACCCCACTAGATGGCGTAGAGTGGCTGCAAATGAAGGATAATGATTTCTCCTATAGACCC ........................................................................................................(((..(((......(((((..((........))..))))).(((....)))..................)))..)))..................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................AAAATGTCCTGACTGAGCTTTCTCCTTCTGTTTCCTTCTTTTTGTCCTGTTT................................................................... | 52 | 1 | 35.00 | 35.00 | 35.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGATGGCGTAGAGTGGCTGCAAATGA.......................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................TAGATGGCGTAGAGTGGCTGCAAATG........................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GGCGTAGAGTGGCTGCAAATGAAGGA...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................CGTAGAGTGGCTGCAAATGAAGGATA.................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................................GATGGCGTAGAGTGGCT.................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................GAGTGGCTGCAAATGAAGGATAATGAT............... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ACAGCTGGCTCGGACAGTGCTACCCTTTTTCATAGGGCACCTTCACGCTGACGTCTGAGTGGTTGTACAATTCACATCCCCACTGGCGGTGTAGGCTCTCATCTGCACCACATTTGTTGTTAGTTCAAGAGAAAATGTCCTGACTGAGCTTTCTCCTTCTGTTTCCTTCTTTTTGTCCTGTTTATCCCCGACCCCACTAGATGGCGTAGAGTGGCTGCAAATGAAGGATAATGATTTCTCCTATAGACCC ........................................................................................................(((..(((......(((((..((........))..))))).(((....)))..................)))..)))..................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................GCTGACGTCTGAGTGGTTGTACAATTCACA.............................................................................................................................................................................. | 30 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - |
| .......GCTCGGACAGTGCTACCCTTTTTCA.......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .................................................GACGTCTGAGTGGTTGTACAATTCACA.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................GCTGACGTCTGAGTGGTTGTACAATTCA................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTCTGTTTCCTTCTTTTTGTCCTGTTTAag.................................................................. | 30 | ag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................CTTCTGTTTCCTTCTTTTTGTCCTGTTTA.................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................CCTTCTGTTTCCTTCTTTTTGTCCTGTTTA.................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................CTTCTGTTTCCTTCTTTTTGTCCTGTT.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |