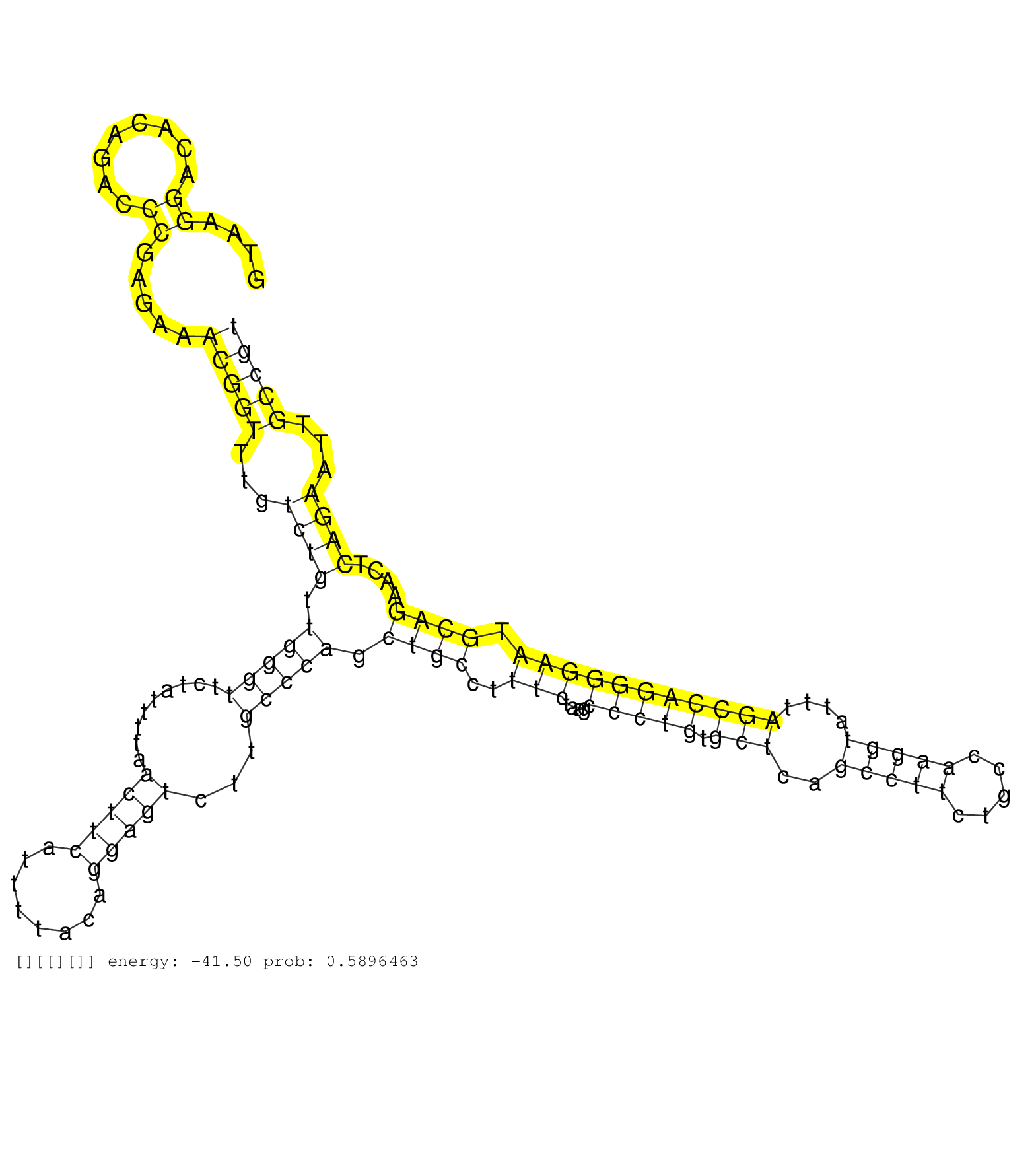

| Gene: Fyn | ID: uc007evx.1_intron_12_0_chr10_39271325_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| TGGAATCTTACTCACAGAGCTGGTCACCAAAGGAAGAGTGCCATACCCAGGTAAGGACACAGACCCGAGAAACGGTTTGTCTGTTGGGTTCTATTTTAACTTCATTTTACAGGAGTCTTGCCCAGCTGCCTTTCTAAGCCCTGTGCTCAGCCTTCTGCCAAGGTATTTAGCCAGGGGAATGCAGAACTCAGAATTGCCGTGACCTGTACACCAAATTCAATGTGCTAAGCAGCTGGTATAAATGTTCCAG ......................................................((........)).....(((((...((((.(((((.........(((((........)))))...))))).((((..(((....(((((.(((..(((((.....)))))....))))))))))).))))....))))...))))).................................................. ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGGACACAGACCCGAGAAACGGTT............................................................................................................................................................................. | 27 | 1 | 20.00 | 20.00 | 9.00 | 4.00 | 3.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - |
| ........TACTCACAGAGCTGGTCACCAAAGGAAGA..................................................................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TTCAATGTGCTAAGCAGCTGGTATAAAT....... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................AGGTATTTAGCCAGGGGAATGCAGAACTC............................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................AGCCAGGGGAATGCAGAACTCAGAATTGC..................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................GCAGAACTCAGAATTGCCGTGA................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TTAGCCAGGGGAATGCAGAACTCAGA.......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GCAGAACTCAGAATTGCCGTGACCTG............................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGGACACAGACCCGAGAAACGGTT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AATGCAGAACTCAGAATTGCCGTGACCTGTA.......................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......TTACTCACAGAGCTGGTCACCAAAGGAA....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................TTTAGCCAGGGGAATGCAGAACTC............................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| TGGAATCTTACTCACAGAGCTGGTCACCAAAGGAAGAGTGCCATACCCAGGTAAGGACACAGACCCGAGAAACGGTTTGTCTGTTGGGTTCTATTTTAACTTCATTTTACAGGAGTCTTGCCCAGCTGCCTTTCTAAGCCCTGTGCTCAGCCTTCTGCCAAGGTATTTAGCCAGGGGAATGCAGAACTCAGAATTGCCGTGACCTGTACACCAAATTCAATGTGCTAAGCAGCTGGTATAAATGTTCCAG ......................................................((........)).....(((((...((((.(((((.........(((((........)))))...))))).((((..(((....(((((.(((..(((((.....)))))....))))))))))).))))....))))...))))).................................................. ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................ACAGACCCGAGAAAacgc.................................................................................................................................................................................. | 18 | acgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |