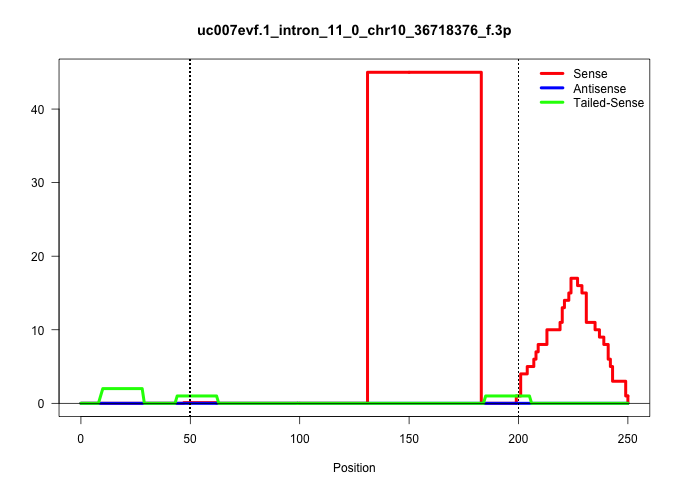
| Gene: Hdac2 | ID: uc007evf.1_intron_11_0_chr10_36718376_f.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| ACCTGAGAGTTTGTTCATTCTTTGGGACCCACATTCTAGAGAACTGTGTCTGCACTGTAAGAAGGAAGTGATCCTAAAGGCAAGAAGCTAAATTATTTTTTTGTTTTTAACATTTATTATATATACACTATATACATACATATATATATTGTGTGTCTTATATTTCAAAATAGACATCAATATTTTTCTTCTGTCTTTAGATGTTAAGGAAGAAGACAAATCCAAGGACAATAGTGGTGAGAAAACAGAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................ATACATACATATATATATTGTGTGTCTTATATTTCAAAATAGACATCAATAT................................................................... | 52 | 1 | 45.00 | 45.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGTTAAGGAAGAAGACAAATCCAAGGACAA................... | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................AGGACAATAGTGGTGAGAA....... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CCAAGGACAATAGTGGTGAGAAAACAGA. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATCCAAGGACAATAGTGGTGAGA........ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TTCTTCTGTCTTTAGATcttc............................................ | 21 | cttc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AAGGACAATAGTGGTGAG......... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TCCAAGGACAATAGTGGTGAGAAAACAGA. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TTTGTTCATTCTTTGGatcc............................................................................................................................................................................................................................. | 20 | atcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................AAGAAGACAAATCCAAGGACAATAGTGG............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................................AGACAAATCCAAGGACAATAGTGGTGAG......... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGTTAAGGAAGAAGACAAATCCAAGG....................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATGTTAAGGAAGAAGACAAATCCAAGGAC..................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................GAAGAAGACAAATCCAAGGACAATAGTGGTG........... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TTGTTCATTCTTTGGaacc............................................................................................................................................................................................................................. | 19 | aacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................................TAAGGAAGAAGACAAATCCAAGGACAA................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TGTGTCTGCACTGTAAcga........................................................................................................................................................................................... | 19 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................................TCCAAGGACAATAGTGGTGAGAAAACAGAC | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGACAAATCCAAGGACAA................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GGAAGAAGACAAATCCAAGGACAATAGT............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................GTCTGCACTGTAAGA............................................................................................................................................................................................ | 15 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 |
| ACCTGAGAGTTTGTTCATTCTTTGGGACCCACATTCTAGAGAACTGTGTCTGCACTGTAAGAAGGAAGTGATCCTAAAGGCAAGAAGCTAAATTATTTTTTTGTTTTTAACATTTATTATATATACACTATATACATACATATATATATTGTGTGTCTTATATTTCAAAATAGACATCAATATTTTTCTTCTGTCTTTAGATGTTAAGGAAGAAGACAAATCCAAGGACAATAGTGGTGAGAAAACAGAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................TCTTCTGTCTTTAGATtca................................................ | 19 | tca | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................CAAAATAGACATCAAtcat...................................................................... | 19 | tcat | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |