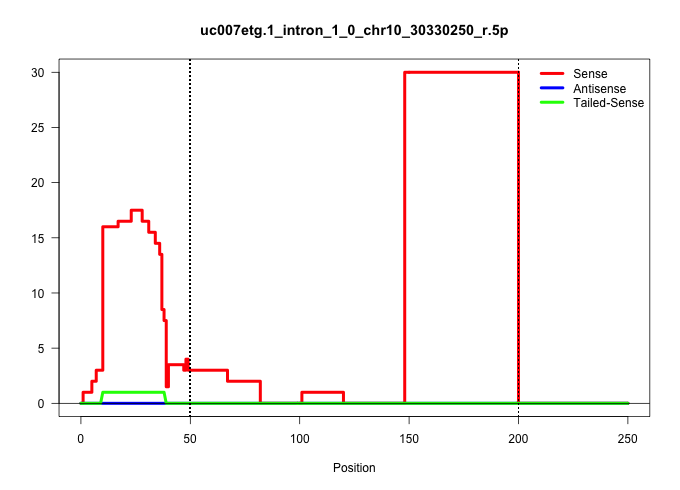
| Gene: Hint3 | ID: uc007etg.1_intron_1_0_chr10_30330250_r.5p | SPECIES: mm9 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| GGGAAAACCATGCTTGAGAGGAATAATTTCACTGACTTCACAGATGTAAGGTATGTACGCTGTGAGTAGACGCTAACACTGGTTCTTGACTTTTCTGAAGTGTGTGTCTCATGAGAAAGGAAGCAGTGGCAGAGTTGTGCATCAGGTGCACCACGTCCTCCTGGACGTGAGGATGCAGTTCATTGATGGTATGAACTTAATGAACCCTGTTTTCATGCCTCACCATCTTCCCTAAACTTCCCTAGTAGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................CACCACGTCCTCCTGGACGTGAGGATGCAGTTCATTGATGGTATGAACTTAA.................................................. | 52 | 1 | 30.00 | 30.00 | 30.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGCTTGAGAGGAATAATTTCACTGACTTC................................................................................................................................................................................................................... | 29 | 1 | 6.00 | 6.00 | - | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........TGCTTGAGAGGAATAATTTCACTGACT..................................................................................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - |
| ........................................CAGATGTAAGGTATGTACGCTGTGAGTAGACGCTAACACTGG........................................................................................................................................................................ | 42 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........TGCTTGAGAGGAATAATTTCACTGACTTt................................................................................................................................................................................................................... | 29 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CCATGCTTGAGAGGAATAATTTCACTG........................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................AGGTATGTACGCTGTGAGT....................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGCTTGAGAGGAATAATTTCACTGAC...................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....AACCATGCTTGAGAGGAATAATTTCA........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGCTTGAGAGGAATAATTTCACTGACTT.................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .GGAAAACCATGCTTGAGAGGAATAATT.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................GTGTGTCTCATGAGAAAGG.................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................TAATTTCACTGACTTCACAGATGTAA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................GAGGAATAATTTCACTGACTTCACAGATGT........................................................................................................................................................................................................... | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| GGGAAAACCATGCTTGAGAGGAATAATTTCACTGACTTCACAGATGTAAGGTATGTACGCTGTGAGTAGACGCTAACACTGGTTCTTGACTTTTCTGAAGTGTGTGTCTCATGAGAAAGGAAGCAGTGGCAGAGTTGTGCATCAGGTGCACCACGTCCTCCTGGACGTGAGGATGCAGTTCATTGATGGTATGAACTTAATGAACCCTGTTTTCATGCCTCACCATCTTCCCTAAACTTCCCTAGTAGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................GAACCCTGTTTTCATGtgt................................. | 19 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |