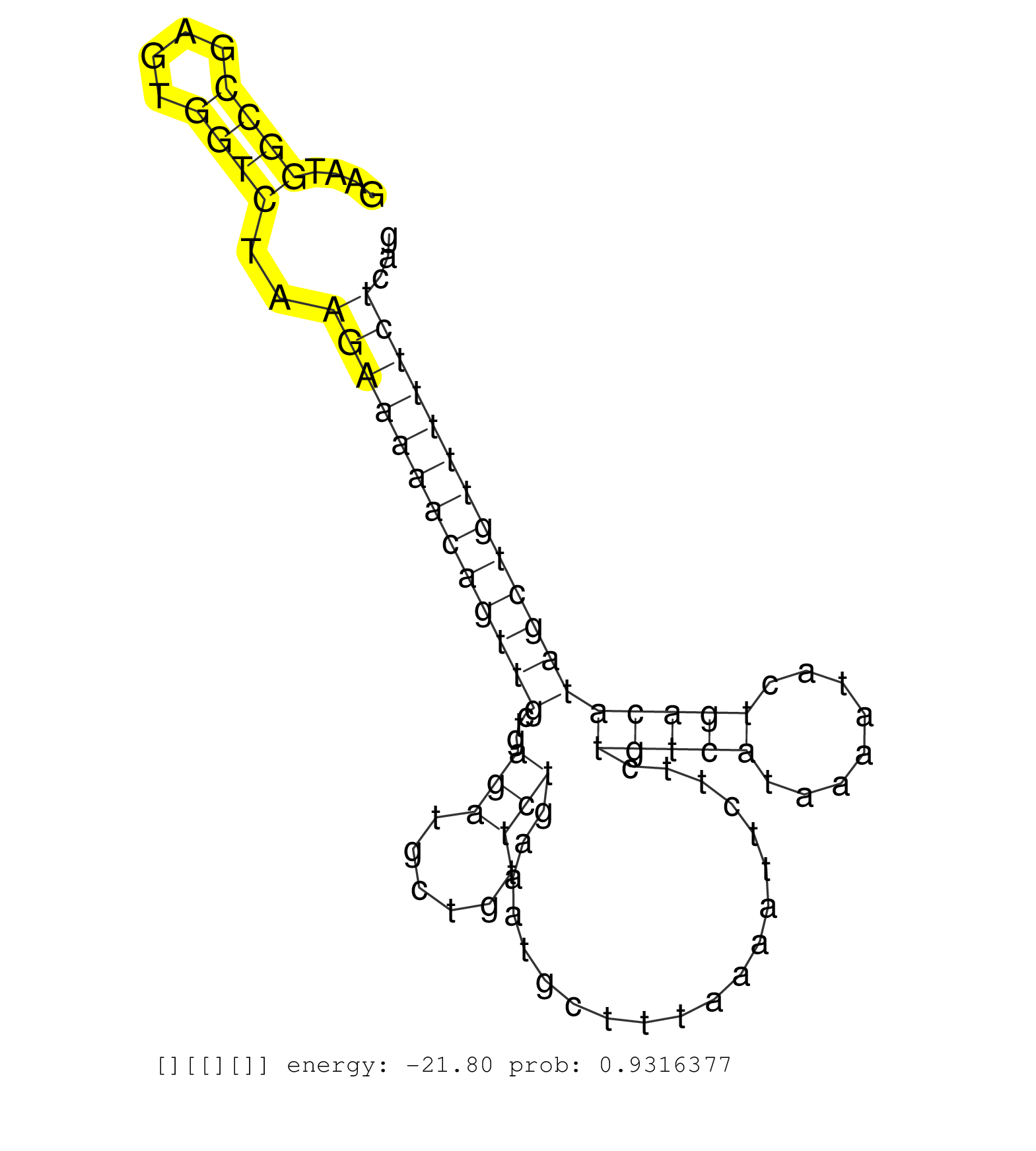

| Gene: L3mbtl3 | ID: uc007esa.1_intron_3_0_chr10_26002666_r.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(4) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(28) TESTES |
| AAAAACCTACCTAAGTGAACATGAAGAAAGTGGGTTCTTAAGTCTGCTGTTGGTGTCAGTTGCTTTAGCCTTTACTTTAAGAAAAACAGTTGCGCCGTCAGAATGGCCGAGTGGTCTAAGAAAAACAGTTGCTGAGATGCTGTTCTGAAATGCTTTAAAATTCTTCTGTCATAAAATACTGACATAGCTGTTTTTCTCAGGGAAAAGCATGCCAATAACTTCAAAGAAGATTCTGAAAAGAAAAAAGAAA ........................................................................................................((((....))))..(((((((((((((...(((......)))....................(((((........))))))))))))))))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGg................................................................................................................................. | 25 | g | 51.00 | 24.00 | 4.00 | 18.00 | 2.00 | 1.00 | 3.00 | 5.00 | - | - | 4.00 | 6.00 | 2.00 | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGgc................................................................................................................................ | 26 | gc | 34.00 | 24.00 | 7.00 | 2.00 | - | 2.00 | 8.00 | 2.00 | 4.00 | 4.00 | - | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAG.................................................................................................................................. | 24 | 1 | 24.00 | 24.00 | 2.00 | 2.00 | - | - | 5.00 | 1.00 | 1.00 | 1.00 | 5.00 | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGG........................................................................................................................................ | 18 | 1 | 12.00 | 12.00 | 3.00 | - | 4.00 | - | - | - | - | 3.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGcgg........................................................................................................................................ | 18 | cgg | 10.33 | 2.83 | 1.83 | - | 1.00 | - | - | 1.00 | 2.67 | 2.00 | - | - | - | - | - | - | 0.17 | 0.17 | - | - | - | - | 0.17 | - | 0.17 | - | - | - | 0.67 | - | 0.33 | - | - | 0.17 |
| ...................................................................................................AGAATGGCCGAGTGGTCTAAG.................................................................................................................................. | 21 | 1 | 10.00 | 10.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGT....................................................................................................................................... | 19 | 1 | 6.00 | 6.00 | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGcggt....................................................................................................................................... | 19 | cggt | 5.83 | 2.83 | 1.00 | - | 0.83 | 0.17 | - | 1.33 | 1.33 | 0.33 | - | - | 0.17 | - | 0.17 | - | - | - | - | - | - | 0.17 | - | 0.17 | - | - | - | - | - | - | - | 0.17 | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCT..................................................................................................................................... | 21 | 1 | 5.00 | 5.00 | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGT.......................................................................................................................................... | 16 | 1 | 5.00 | 5.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTA.................................................................................................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTC...................................................................................................................................... | 20 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGAATGGCCGAGTGGTCTA.................................................................................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAA................................................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTG......................................................................................................................................... | 17 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGAGTGGTCTAAGgctg.............................................................................................................................. | 17 | gctg | 4.00 | 0.00 | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAG........................................................................................................................................... | 15 | 6 | 2.83 | 2.83 | 1.33 | - | - | - | - | 0.33 | 0.67 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGgccc.............................................................................................................................. | 28 | gccc | 2.00 | 24.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGcg......................................................................................................................................... | 17 | cg | 2.00 | 2.83 | 0.50 | - | 0.50 | - | - | 0.67 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGgcgc.............................................................................................................................. | 28 | gcgc | 2.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGAATGGCCGAGTGGTCT..................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGc.......................................................................................................................................... | 16 | c | 1.33 | 2.83 | 0.67 | - | - | - | - | 0.17 | 0.33 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGAATGGCCGAGTGGTCTAAGgcgc.............................................................................................................................. | 25 | gcgc | 1.00 | 10.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGAGTGGTCTAtggc................................................................................................................................ | 15 | tggc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAtt................................................................................................................................. | 25 | tt | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGaat...................................................................................................................................... | 20 | aat | 1.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATGGCCGAGTGGTCTcagg................................................................................................................................. | 19 | cagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CAGAATGGCCGAGTGGTCTAAG.................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGgca............................................................................................................................... | 27 | gca | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CAGAATGGCCGAGTGGTCT..................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGtc................................................................................................................................ | 26 | tc | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGAATGGCCGAGTGGTCTAtgc................................................................................................................................. | 22 | tgc | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGAGTGGTCTAAGgcga.............................................................................................................................. | 17 | gcga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGgcg............................................................................................................................... | 27 | gcg | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGga................................................................................................................................ | 26 | ga | 1.00 | 24.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAGgcc............................................................................................................................... | 27 | gcc | 1.00 | 24.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAcg................................................................................................................................. | 25 | cg | 1.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCa..................................................................................................................................... | 21 | a | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGTGGTCTAAtgc................................................................................................................................ | 26 | tgc | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GCGCCGTCAGAATGGCCGAGTGGTCTAAGA................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................ATGGCCGAGTGGTCTAAG.................................................................................................................................. | 18 | 9 | 0.89 | 0.89 | - | - | - | 0.44 | 0.11 | - | - | - | - | - | - | - | - | 0.22 | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGaa......................................................................................................................................... | 17 | aa | 0.50 | 2.83 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TTAAGTCTGCTGTTGG..................................................................................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGcga........................................................................................................................................ | 18 | cga | 0.33 | 2.83 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGcggc....................................................................................................................................... | 19 | cggc | 0.17 | 2.83 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GTCAGAATGGCCGAGcgga....................................................................................................................................... | 19 | cgga | 0.17 | 2.83 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - |
| ......................................................................................................ATGGCCGAGTGGTCTAAGgcc............................................................................................................................... | 21 | gcc | 0.11 | 0.89 | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAAAACCTACCTAAGTGAACATGAAGAAAGTGGGTTCTTAAGTCTGCTGTTGGTGTCAGTTGCTTTAGCCTTTACTTTAAGAAAAACAGTTGCGCCGTCAGAATGGCCGAGTGGTCTAAGAAAAACAGTTGCTGAGATGCTGTTCTGAAATGCTTTAAAATTCTTCTGTCATAAAATACTGACATAGCTGTTTTTCTCAGGGAAAAGCATGCCAATAACTTCAAAGAAGATTCTGAAAAGAAAAAAGAAA ........................................................................................................((((....))))..(((((((((((((...(((......)))....................(((((........))))))))))))))))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | GSM475281(GSM475281) total RNA. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|