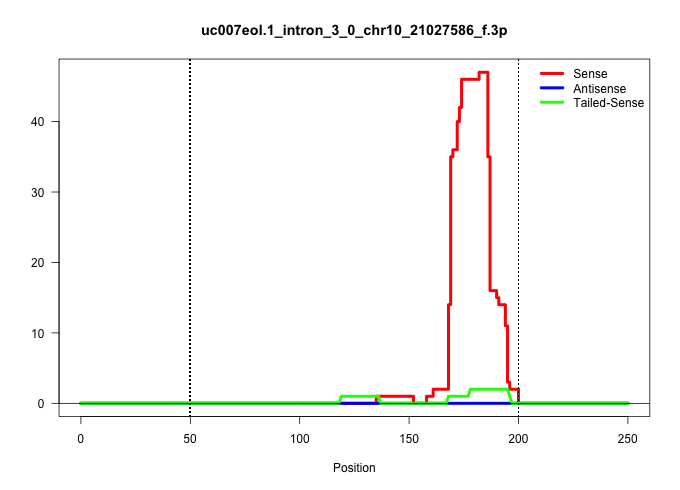
| Gene: Hbs1l | ID: uc007eol.1_intron_3_0_chr10_21027586_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| AGGTTGTTAGAGGTAATACTCCTTTTTTATTTTCCTCATGGTCCCTATTGCTAAGTAATAAACAAGTATCATTTATATCATGATCAAGAGATGCACTTAGTAATTAAATGCCTCGTTTCATGTCTCCTTTTGGTTGCTGTGACTGTCTCCTGCAAATGTCAACTCATGCATCTGAACATTGATGGCTCTTGTAAGGTAAGGAGTCTTATTTTCTTCCTTTGGAGTTTCACCCCAAAATGTTCACCATTCT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................ATCTGAACATTGATGGCT............................................................... | 18 | 1 | 19.00 | 19.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CATCTGAACATTGATGGC................................................................ | 18 | 1 | 11.00 | 11.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AACATTGATGGCTCTTGTAAG....................................................... | 21 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGAACATTGATGGCTCTTGTAAG....................................................... | 23 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................GAACATTGATGGCTCTTGTAA........................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................CATCTGAACATTGATGGCTCTTG........................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CATCTGAACATTGATGGCTCTTGTAAGGa..................................................... | 29 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................ATCTGAACATTGATGGCTCTTGTAA........................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGAACATTGATGGCTCTTGTAAGGTAAG.................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGGCTCTTGTAAGGTAAG.................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................TTGATGGCTCTTGTtcta...................................................... | 18 | tcta | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TCAACTCATGCATCTGAACATTGATGGC................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACTCATGCATCTGAACATTGATGGCTCTT............................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GCTGTGACTGTCTCCTG.................................................................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................ATCTGAACATTGATGGCTCTTGTAAG....................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................TCTGAACATTGATGGCTCTTGTAAGG...................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................ATGTCTCCTTTTGGgtat................................................................................................................. | 18 | gtat | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGTTGTTAGAGGTAATACTCCTTTTTTATTTTCCTCATGGTCCCTATTGCTAAGTAATAAACAAGTATCATTTATATCATGATCAAGAGATGCACTTAGTAATTAAATGCCTCGTTTCATGTCTCCTTTTGGTTGCTGTGACTGTCTCCTGCAAATGTCAACTCATGCATCTGAACATTGATGGCTCTTGTAAGGTAAGGAGTCTTATTTTCTTCCTTTGGAGTTTCACCCCAAAATGTTCACCATTCT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................TGTCTCCTGCAAATtggt............................................................................................. | 18 | tggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................................GAGTCTTATTTTCTTaggg................................... | 19 | aggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |